Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) - PubMed (original) (raw)
Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L)
Weiliang Fan et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The class III phosphatidylinositol 3-kinase (PI3KC3) is crucial for autophagosome biogenesis. It has been long speculated to nucleate the autophagosome membrane, but the biochemical mechanism of such nucleation activity remains unsolved. We recently identified Barkor/Atg14(L) as the targeting factor for PI3KC3 to autophagosome membrane. Here, we show that we have characterized the region of Barkor/Atg14(L) required for autophagosome targeting and identified the BATS [Barkor/Atg14(L) autophagosome targeting sequence] domain at the carboxyl terminus of Barkor. Bioinformatics and mutagenesis analyses revealed that the BATS domain binds to autophagosome membrane via the hydrophobic surface of an intrinsic amphipathic alpha helix. BATS puncta overlap with Atg16 and LC3, and partially with DFCP1, in a stress-inducible manner. Ectopically expressed BATS accumulates on highly curved tubules that likely represent intermediate autophagic structures. PI3KC3 recruitment and autophagy stimulation by Barkor/Atg14(L) require the BATS domain. Furthermore, our biochemical analyses indicate that the BATS domain directly binds to the membrane, and it favors membrane composed of phosphatidylinositol 3-phosphate [PtdIns(3)P] and phosphatidylinositol 4,5-biphosphate [PtdIns(4,5)P2]. By binding preferentially to curved membranes incorporated with PtdIns(3)P but not PtdIns(4,5)P2, the BATS domain is capable of sensing membrane curvature. Thus, we propose a novel model of PI3KC3 autophagosome membrane nucleation in which its autophagosome-specific adaptor, Barkor, accumulates on highly curved PtdIns(3)P enriched autophagic membrane via its BATS domain to sense and maintain membrane curvature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
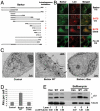
The identification of the BATS domain required for autophagosome targeting and autophagy activation. (A). Schematic representation of the deletion mutants of Barkor. All mutants are tagged with hrGFP and FLAG. Autophagosome localization is defined as cytosolic puncta overlapping with LC3 upon CQ treatment. (B). Cells stably expressing Myc-LC3 were transfected with various Barkor/Atg14(L) fragments tagged with hrGFP. Cells were treated with 200 μM CQ for 2 h and stained with anti–c-Myc antibody for LC3 and green fluorescence for Barkor/Atg14(L) mutants. (C). U2OS cells that overexpress full-length Barkor, BarkorΔ10aa and U2OS parental cells were visualized using a transmission electron microscope. AVs are marked by black arrows. Scale bar, 2 μm. (D). AVs per cross-section were counted (control, 3.45 ± 0.35; Barkor/Atg14(L) Δ10aa, 3.5 ± 0.32; Barkor/Atg14(L) WT: 12.6 ± 0.9). (E). LC3 and tubulin were detected in cells described in C; 2-h treatment with 50 nM Bafilomycin was used to block lysosomal degradation.
Fig. 2.
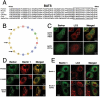
The amphipathic alpha helix of BATS is responsible for autophagosome membrane binding. (A). The last 80 amino acids (BATS) of Barkor/Atg14(L) homologues in vertebrates were aligned, and a highly conserved membrane interaction helix was predicted and noted by a box. (B). The predicted membrane interaction helix is presented as an amphipathic helical wheel. Hydrophobic residues are yellow, the hydrophilic residues are blue, and positively charged residues are green. Three hydrophobic residues mutated in C are marked by arrows. (C). Cells stably expressing Myc-LC3 were coexpressed with different BATS mutants, which were tagged with hrGFP (BATS K486A, R492A and BATS WFYm-BATS mutant W484R, F485R, Y488R). Cells were treated with 200 μM CQ for 2 h and stained with c-Myc antibody for LC3 and green fluorescence for BATS mutants. (D). U2OS cells were coexpressed with Myc-Beclin 1 and different Barkor/Atg14(L) mutants tagged by hrGFP and treated with 200 μM CQ. (E). Cells stably expressing Myc-LC3 were coexpressed with either GFP-Beclin 1 or GFP-Beclin 1-BATS, and treated with 200 μM CQ for 2 h.
Fig. 3.
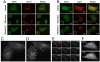
BATS localizes to highly curved early autophagic structures. (A). U2OS cell were transfected with hrGFP-BATS together with Myc-DFCP1. Forty-eight hours after transfection, cells were either treated by EBSS for 1 h or by 200 μM CQ for 2 h; Myc-DFCP1 was stained with Rhodamine Red antibody and detected by laser confocal microscope. (B) U2OS cells were transfected with hrGFP-BATS together with Myc-Atg16. Forty-eight hours after transfection, cells were either treated with EBSS for 1 h or with 2 μM rapamycin for 4 h; Myc-Atg16 was stained with Rhodamine Red antibody and detected by laser confocal microscope. (C and D) U2OS cells expressing GFP-tagged BATS (2 d after transfection) were treated with EBSS medium and observed under the fluorescence microscope. Images were captured immediately after the addition of EBSS medium (C) or at 30-min time points (D) (also see
Movie S1
). Arrows mark tubules. (E). Arrows along the time course (0–30 min) mark one representative tubule after starvation. (F). U2OS cells expressing GFP-BATS and Tomato-LC3 were captured after addition of EBSS medium; both channels were captured every 2 s. Two representative images were presented (also see
Movie S2
). An arrow marks a tubule.
Fig. 4.
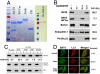
Membrane binding activity of the BATS domain. (A). GST-tagged constructs of wild-type BATS, BATS WFYm mutant, endophilin 1, and PH domain of PLCδ were recombinantly expressed and purified from E.coli. Full-length His-tagged Barkor/Atg14(L) was expressed and purified from baculovirus-infected insect cells. (B). Purified recombinant proteins described in A were tested for their liposome binding activity in a cosedimentation assay. Liposomes generated from Folch fraction I of brain extract from bovine brains were used in this assay. S denotes supernatant and P denotes pellet. The binding efficiency is calculated as bound proteins (P) versus input (P + S). (C). Recombinant wild-type BATS, BATS WFYm mutant and the PH domain of PLCδ were incubated with liposomes incorporated with different forms of phosphoinositides and tested for their liposome binding in a cosedimentation assay. S denotes supernatant and P denotes pellet. (D). Cells expressing GFP-BATS and Myc-LC3 were left untreated, treated with Rapamycin alone or in combination with 3-Methyladenine (3-MA), and observed under the fluorescence microscope. Dots per cells were calculated in at least 50 cells from three independent experiments.
Fig. 5.
Membrane curvature sensing by the BATS domain. (A). Folch fraction derived liposomes with different sizes (100–800 nm) were incubated with recombinant BATS in a cosedimentation assay. S denotes supernatant and P denotes pellet. (B). Liposome binding was evaluated as the percentage of BATS bound to liposomes compared to the binding of BATS to 800 nm liposomes. Liposome loading was evaluated by the measurement of fluorescence-labeled PE in the liposomes. (C). Three hundred fifty micromolar lipid with equal amount of PC, PE combining a gradient increased ratio of PtdIns(3)P (0.2, 0.6, 2, and 6 mol%) were made into liposomes with two different sizes (100 and 800 nm), and their binding to recombinant BATS were tested by cosedimentation assays. S denotes supernatant and P denotes pellet. The binding efficiency (bound protein versus input) is denoted as P/(P + S). (D). The similar liposome binding assay as described in (C) was performed using PtdIns(4,5)P2. (E). Proposed functional model of Barkor/Atg14(L) BATS domain in the membrane targeting of Barkor/Atg14(L) on highly curved PtdIns(3)P enriched membrane. N, N terminus; C, C terminus. Please see text for more explanation.
Similar articles
- Membrane curvature response in autophagy.
Wilz L, Fan W, Zhong Q. Wilz L, et al. Autophagy. 2011 Oct;7(10):1249-50. doi: 10.4161/auto.7.10.16738. Epub 2011 Oct 1. Autophagy. 2011. PMID: 21738011 Free PMC article. - PtdIns(4,5)P2 signaling regulates ATG14 and autophagy.
Tan X, Thapa N, Liao Y, Choi S, Anderson RA. Tan X, et al. Proc Natl Acad Sci U S A. 2016 Sep 27;113(39):10896-901. doi: 10.1073/pnas.1523145113. Epub 2016 Sep 12. Proc Natl Acad Sci U S A. 2016. PMID: 27621469 Free PMC article. - ATG14 controls SNARE-mediated autophagosome fusion with a lysosome.
Liu R, Zhi X, Zhong Q. Liu R, et al. Autophagy. 2015;11(5):847-9. doi: 10.1080/15548627.2015.1037549. Autophagy. 2015. PMID: 25945523 Free PMC article. - WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome.
Proikas-Cezanne T, Takacs Z, Dönnes P, Kohlbacher O. Proikas-Cezanne T, et al. J Cell Sci. 2015 Jan 15;128(2):207-17. doi: 10.1242/jcs.146258. J Cell Sci. 2015. PMID: 25568150 Review. - Membrane targeting of core autophagy players during autophagosome biogenesis.
Dudley LJ, Makar AN, Gammoh N. Dudley LJ, et al. FEBS J. 2020 Nov;287(22):4806-4821. doi: 10.1111/febs.15334. Epub 2020 May 1. FEBS J. 2020. PMID: 32301577 Review.
Cited by
- Autophagy and its role in regeneration and remodeling within invertebrate.
Song Q, Liu H, Zhen H, Zhao B. Song Q, et al. Cell Biosci. 2020 Sep 21;10:111. doi: 10.1186/s13578-020-00467-3. eCollection 2020. Cell Biosci. 2020. PMID: 32974004 Free PMC article. Review. - Targeting PI3K family with small-molecule inhibitors in cancer therapy: current clinical status and future directions.
Li H, Wen X, Ren Y, Fan Z, Zhang J, He G, Fu L. Li H, et al. Mol Cancer. 2024 Aug 10;23(1):164. doi: 10.1186/s12943-024-02072-1. Mol Cancer. 2024. PMID: 39127670 Free PMC article. Review. - Sensing Membrane Curvature in Macroautophagy.
Nguyen N, Shteyn V, Melia TJ. Nguyen N, et al. J Mol Biol. 2017 Feb 17;429(4):457-472. doi: 10.1016/j.jmb.2017.01.006. Epub 2017 Jan 11. J Mol Biol. 2017. PMID: 28088480 Free PMC article. Review. - Anatomy of autophagy: from the beginning to the end.
Zhi X, Feng W, Rong Y, Liu R. Zhi X, et al. Cell Mol Life Sci. 2018 Mar;75(5):815-831. doi: 10.1007/s00018-017-2657-z. Epub 2017 Sep 22. Cell Mol Life Sci. 2018. PMID: 28939950 Free PMC article. Review. - PAQR3 controls autophagy by integrating AMPK signaling to enhance ATG14L-associated PI3K activity.
Xu DQ, Wang Z, Wang CY, Zhang DY, Wan HD, Zhao ZL, Gu J, Zhang YX, Li ZG, Man KY, Pan Y, Wang ZF, Ke ZJ, Liu ZX, Liao LJ, Chen Y. Xu DQ, et al. EMBO J. 2016 Mar 1;35(5):496-514. doi: 10.15252/embj.201592864. Epub 2016 Feb 1. EMBO J. 2016. PMID: 26834238 Free PMC article.
References
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. - PubMed
- Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
- Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials