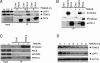TWEAK induces apoptosis through a death-signaling complex comprising receptor-interacting protein 1 (RIP1), Fas-associated death domain (FADD), and caspase-8 - PubMed (original) (raw)
TWEAK induces apoptosis through a death-signaling complex comprising receptor-interacting protein 1 (RIP1), Fas-associated death domain (FADD), and caspase-8
Aminah Ikner et al. J Biol Chem. 2011.
Abstract
The tumor necrosis factor (TNF) superfamily member TNF-like weak inducer of apoptosis (TNFSF12, CD255) (TWEAK) can stimulate apoptosis in certain cancer cells. Previous studies suggest that TWEAK activates cell death indirectly, by inducing TNFα-mediated autocrine signals. However, the underlying death-signaling mechanism has not been directly defined. Consistent with earlier work, TWEAK assembled a proximal signaling complex containing its cognate receptor FN14, the adaptor TRAF2, and cellular inhibitor of apoptosis protein 1 (cIAP1). Neither the death domain adaptor Fas-associated death domain nor the apoptosis-initiating protease caspase-8 associated with this primary complex. Rather, TWEAK induced TNFα secretion and TNF receptor 1-dependent assembly of a death-signaling complex containing receptor-interacting protein 1 (RIP1), FADD, and caspase-8. Knockdown of RIP1 by siRNA prevented TWEAK-induced association of FADD with caspase-8 but not formation of the FN14-TRAF2-cIAP1 complex and inhibited apoptosis activation. Depletion of the RIP1 E3 ubiquitin ligase cIAP1 enhanced assembly of the RIP1-FADD-caspase-8 complex and augmented cell death. Conversely, knockdown of the RIP1 deubiquitinase CYLD inhibited these functions. Depletion of FADD, caspase-8, BID, or BAX and BAK but not RIP3 attenuated TWEAK-induced cell death. Pharmacologic inhibition of the NF-κB pathway or siRNA knockdown of RelA attenuated TWEAK induction of TNFα and association of RIP1 with FADD and caspase-8. These results suggest that TWEAK triggers apoptosis by promoting assembly of a RIP1-FADD-caspse-8 complex via autocrine TNFα-TNFR1 signaling. The proapoptotic activity of TWEAK is modulated by cIAP1 and CYLD and engages both the extrinsic and intrinsic signaling pathways.
Figures
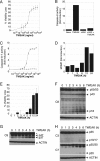
FIGURE 1.
TWEAK activates apoptosis in HSC3 cells. A, cells were treated with the indicated concentrations of TWEAK for 48 h and assayed for viability. B, after cells were treated with TWEAK (3 μg/ml, 6 h), caspase-8 was subjected to IP and assayed for activity. C, cells were treated with the indicated concentrations of TWEAK for 48 h in the absence (●) or presence (○) of the pan-caspase inhibitor zVAD-FMK (20 μ
m
) and assayed for caspase-3/7 activity. D, cells were treated with TWEAK (3 μg/ml) for the indicated number of hours and assayed for MOMP or viability (E). F–H, cells were treated with TWEAK (10 μg/ml) for the indicated number of hours and subjected to IB analysis with antibodies specific to caspase-8 (F), caspase-9 (G), or caspase-3 and caspase-7 (H). The asterisk indicates a nonspecific band.
FIGURE 2.
TWEAK assembles a proximal signaling complex containing FN14, TRAF2, and cIAP1. A, cells were treated with TWEAK (3 μg/ml) for the indicated amount of time and subjected to IP of FN14 followed by IB with antibodies to FN14, TRAF2, and cIAP1. B, cells were treated with TWEAK (3 μg/ml, 0.5 h) and subjected to IP of FN14 or TRAF2, followed by IB analysis of FN14 and TRAF2. C, cells were treated with TWEAK (3 μg/ml, 0.5 h) and subjected to IP of TRAF2 followed by IB of FN14, FADD, caspase-8, or TRAF2. D, cells were treated with TWEAK (3 μg/ml) for the indicated time and subjected to IB analysis of TRAF2 and cIAP1. The asterisk indicates a nonspecific band.
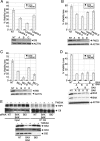
FIGURE 3.
TWEAK induces association of a death-signaling complex containing RIP1, FADD, and caspase-8, leading to caspase-8 activation. A, cells were treated with TWEAK (3 μg/ml) for the indicated time in the absence (D-dimethyl sulfoxide) or presence of zVAD-FMK and subjected to IP of caspase-8 followed by IB analysis of caspase-8 and RIP1. B, cells were treated with TWEAK (3 μg/ml, 6 h) and subjected to IP of RIP1 followed by IB analysis of FN14, FADD, caspase-8, or RIP1. C, cells were treated with TWEAK (3 μg/ml, 6 h), RIP1 was immunoprecipitated from cell lysates, and IPs were assayed for caspase-8 activity. D, cells were transfected with two siRNA oligonucleotides against RIP1 for 48 h, treated with TWEAK (3 μg/ml, 6 h) and subjected to IP of caspase-8 followed by IB analysis of FADD or caspase-8. E, cells were transfected with siRNA against FADD for 48 h, treated with TWEAK (3 μg/ml, 6 h), and subjected to IP of caspase-8 followed by IB analysis of FADD, caspase-8, or RIP1. F–H, cells were transfected with RIP1 siRNA as in D, treated with TWEAK for 48 h, and then analyzed for viability (F), caspase-3/7 activity (G), or MOMP (H). In F, IB analysis shows siRNA knockdown levels. For the IP experiments, zVAD-FMK (20 μ
m
) was added 1 h prior to incubation with TWEAK. A vertical line denotes lane removal from the original image.
FIGURE 4.
TWEAK induces TNFα and inhibiting NF-κB abrogates formation of the proapoptotic complex between RIP1-FADD-C8. A, cells were incubated with TWEAK (3 μg/ml) for the indicated amount of time, and cell supernatants were analyzed with a TNFα-specific ELISA. B, cells were transfected with FN14 siRNA for 48 h, incubated with TWEAK (3 μg/ml) for 48 h, and analyzed for TNFα secretion as in A. C, HSC3 cells were treated with BMS-345541 (30 μ
m
) in the absence or presence of TWEAK (3 μg/ml) for 24 h. Cell supernatants were analyzed with a TNFα-specific ELISA. D, cells were treated with TWEAK (3 μg/ml) for 6 h in the presence of zVAD-FMK and BMS-345541. Subsequently, caspase-8 was immunoprecipitated from whole cell lysates followed by IP of RIP1, FADD, and caspase-8. E, HSC3 cells were transfected with two siRNA oligonucleotides against RelA for 48 h and then treated with TWEAK (3 μg/ml, 24 h). Cell supernatants were analyzed with a TNFα-specific ELISA. F, cells transfected with RelA siRNA were treated with TWEAK (3 μg/ml) for 6 h in presence of zVAD-FMK (20 μ
m
), and caspase-8 was immunoprecipitated followed by IB analysis of RIP1, FADD, caspase-8, and RelA.
FIGURE 5.
TWEAK induces TNFα and drives TNFR1-dependent apoptosis signaling. A, cells were transfected with TNFR1 siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by immunoblot analysis as indicated. B, cells were transfected with TNFR1 siRNA and treated with TWEAK as in A, and assembly of the FN14-TRAF2-cIAP1 complex was examined by FN14 IP followed by immunoblot analysis as indicated. C–E, cells were transfected with TNFR1 siRNA, treated with TWEAK as in A, and analyzed for viability (C), caspase-3/7 activity (D), and MOMP (E). In B, IB analysis shows siRNA knockdown levels. For the IP experiments, zVAD-FMK (20 μ
m
) was added 1 h prior to incubation with TWEAK. F, HSC3 cells were treated with TWEAK (3 μg/ml) for 6 h in the presence of zVAD-FMK (20 μ
m
) and TNFR1:FC (100 μg/ml). Cell lysates were immunoprecipitated with caspase-8 followed by IB analysis of RIP1, FADD, and caspase-8.
FIGURE 6.
Modulation of TWEAK-induced apoptosis by cIAP1 and CYLD. A, cells were transfected with cIAP1 siRNA for 48 h and stimulated with TWEAK (3 μg/ml, 6 h), and association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by IB analysis of FADD, caspase-8, cIAP1, or RIP1. B, cells were transfected with cIAP1 siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 48 h), and then analyzed for viability. IB analysis shows siRNA knockdown levels. C, cells were transfected with CYLD siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and then association of RIP1 and caspase-8 was analyzed by caspase-8 IP followed by IB analysis of FADD, caspase-8, or RIP1. D, cells were transfected with CYLD siRNA for 48 h, stimulated with TWEAK (3 μg/ml, 48 h), and then analyzed for viability. IB analysis shows siRNA knockdown levels.
FIGURE 7.
TWEAK requires both the extrinsic and intrinsic apoptosis signaling pathways to induce cell death. A–D, cells were transfected with siRNAs oligonucleotides against caspase-8 (A), FADD (B), BID (C), or BAX and BAK (D) for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and then analyzed for cell viability. In A–D, IB analysis shows siRNA knockdown levels. E, cells were transfected with siRNA against BAX and BAK or BID for 48 h, stimulated with TWEAK (3 μg/ml, 6 h), and subjected to caspase-8 IP followed by IB analysis of BID, BAX, BAK, caspase-8, or RIP1.
Similar articles
- RIP1 is required for IAP inhibitor-mediated sensitization for TRAIL-induced apoptosis via a RIP1/FADD/caspase-8 cell death complex.
Abhari BA, Cristofanon S, Kappler R, von Schweinitz D, Humphreys R, Fulda S. Abhari BA, et al. Oncogene. 2013 Jul 4;32(27):3263-73. doi: 10.1038/onc.2012.337. Epub 2012 Aug 13. Oncogene. 2013. PMID: 22890322 - TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1-TRAF2 complex to sensitize tumor cells to TNFalpha.
Vince JE, Chau D, Callus B, Wong WW, Hawkins CJ, Schneider P, McKinlay M, Benetatos CA, Condon SM, Chunduru SK, Yeoh G, Brink R, Vaux DL, Silke J. Vince JE, et al. J Cell Biol. 2008 Jul 14;182(1):171-84. doi: 10.1083/jcb.200801010. Epub 2008 Jul 7. J Cell Biol. 2008. PMID: 18606850 Free PMC article. - The adaptor protein FADD and the initiator caspase-8 mediate activation of NF-κB by TRAIL.
Grunert M, Gottschalk K, Kapahnke J, Gündisch S, Kieser A, Jeremias I. Grunert M, et al. Cell Death Dis. 2012 Oct 25;3(10):e414. doi: 10.1038/cddis.2012.154. Cell Death Dis. 2012. PMID: 23096115 Free PMC article. - Characterization of the ripoptosome and its components: implications for anti-inflammatory and cancer therapy.
Schilling R, Geserick P, Leverkus M. Schilling R, et al. Methods Enzymol. 2014;545:83-102. doi: 10.1016/B978-0-12-801430-1.00004-4. Methods Enzymol. 2014. PMID: 25065887 Review. - The roles of FADD in extrinsic apoptosis and necroptosis.
Lee EW, Seo J, Jeong M, Lee S, Song J. Lee EW, et al. BMB Rep. 2012 Sep;45(9):496-508. doi: 10.5483/bmbrep.2012.45.9.186. BMB Rep. 2012. PMID: 23010170 Review.
Cited by
- Intermediate domain of receptor-interacting protein kinase 1 (RIPK1) determines switch between necroptosis and RIPK1 kinase-dependent apoptosis.
Duprez L, Bertrand MJ, Vanden Berghe T, Dondelinger Y, Festjens N, Vandenabeele P. Duprez L, et al. J Biol Chem. 2012 Apr 27;287(18):14863-72. doi: 10.1074/jbc.M111.288670. Epub 2012 Feb 23. J Biol Chem. 2012. PMID: 22362767 Free PMC article. - Licochalcone-A induces intrinsic and extrinsic apoptosis via ERK1/2 and p38 phosphorylation-mediated TRAIL expression in head and neck squamous carcinoma FaDu cells.
Park MR, Kim SG, Cho IA, Oh D, Kang KR, Lee SY, Moon SM, Cho SS, Yoon G, Kim CS, Oh JS, You JS, Kim DK, Seo YS, Im HJ, Kim JS. Park MR, et al. Food Chem Toxicol. 2015 Mar;77:34-43. doi: 10.1016/j.fct.2014.12.013. Epub 2015 Jan 5. Food Chem Toxicol. 2015. PMID: 25572524 Free PMC article. - Tumor Necrosis Factor (TNF) Receptor Expression Determines Keratinocyte Fate upon Stimulation with TNF-Like Weak Inducer of Apoptosis.
Wang X, Cheng D, Hu G, Liang L, Tan F, Xiao T, Xiao S, Xia Y. Wang X, et al. Mediators Inflamm. 2019 Dec 5;2019:2945083. doi: 10.1155/2019/2945083. eCollection 2019. Mediators Inflamm. 2019. PMID: 31885495 Free PMC article. - Exocyst inactivation in urothelial cells disrupts autophagy and activates non-canonical NF-κB signaling.
Ortega MA, Villiger RK, Harrison-Chau M, Lieu S, Tamashiro KK, Lee AJ, Fujimoto BA, Patwardhan GY, Kepler J, Fogelgren B. Ortega MA, et al. Dis Model Mech. 2022 Oct 1;15(10):dmm049785. doi: 10.1242/dmm.049785. Epub 2022 Oct 12. Dis Model Mech. 2022. PMID: 36004645 Free PMC article. - HPV Type 16 Infection Switches Keratinocytes from Apoptotic to Proliferative Fate under TWEAK/Fn14 Interaction.
Cheng H, Zhan N, Ding D, Liu X, Zou X, Li K, Xia Y. Cheng H, et al. J Invest Dermatol. 2015 Oct;135(10):2427-2436. doi: 10.1038/jid.2015.201. Epub 2015 May 27. J Invest Dermatol. 2015. PMID: 26016896
References
- Chicheportiche Y., Bourdon P. R., Xu H., Hsu Y. M., Scott H., Hession C., Garcia I., Browning J. L. (1997) J. Biol. Chem. 272, 32401–32410 - PubMed
- Marsters S. A., Sheridan J. P., Pitti R. M., Brush J., Goddard A., Ashkenazi A. (1998) Curr. Biol. 8, 525–528 - PubMed
- Vince J. E., Silke J. (2006) Cell Death Differ. 13, 1842–1844 - PubMed
- Gao H. X., Campbell S. R., Burkly L. C., Jakubowski A., Jarchum I., Banas B., Saleem M. A., Mathieson P. W., Berman J. W., Michaelson J. S., Putterman C. (2009) Cytokine 46, 24–35 - PubMed
- Burkly L. C., Michaelson J. S., Hahm K., Jakubowski A., Zheng T. S. (2007) Cytokine 40, 1–16 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous