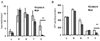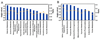The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon - PubMed (original) (raw)
The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon
Dallas R Donohoe et al. Cell Metab. 2011.
Abstract
The microbiome is being characterized by large-scale sequencing efforts, yet it is not known whether it regulates host metabolism in a general versus tissue-specific manner or which bacterial metabolites are important. Here, we demonstrate that microbiota have a strong effect on energy homeostasis in the colon compared to other tissues. This tissue specificity is due to colonocytes utilizing bacterially produced butyrate as their primary energy source. Colonocytes from germfree mice are in an energy-deprived state and exhibit decreased expression of enzymes that catalyze key steps in intermediary metabolism including the TCA cycle. Consequently, there is a marked decrease in NADH/NAD(+), oxidative phosphorylation, and ATP levels, which results in AMPK activation, p27(kip1) phosphorylation, and autophagy. When butyrate is added to germfree colonocytes, it rescues their deficit in mitochondrial respiration and prevents them from undergoing autophagy. The mechanism is due to butyrate acting as an energy source rather than as an HDAC inhibitor.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Figure 1. Effects of Microbiota on Energy Homeostasis
Total NADH/NAD+ ratios (A) and ATP levels (B) in various tissues from GF and CONV-R mice. L, liver; H, heart; K, kidney; T, testis; C, colon. 6 GF and 6 CONV-R mice were analyzed, and results are mean ± SEM. Significant differences are indicated (**p <0.01).
Figure 2. Analysis of Microbiome Regulated Gene Products and Metabolites
Top KEGG pathways of transcriptome (A) and proteome (B) identified as differences between GF and CONV-R. Blue bars correspond to −log (p-values) shown on y-axis at left. Straight yellow lines represent threshold of significance for these −log (p-values). Yellow boxes represent the ratio, shown on y-axis at right, and indicate the number of genes or proteins affected divided by total number within category. 6 GF and 6 CONV-R mice were used for the transcriptome analysis. For the proteome analysis, 3 GF and 3 CONV-R mice were used.
Figure 3. Regulation of Colonocyte Oxidative Metabolism
(A) Percentage of 13C-butyrate metabolized to 13CO2 in CONV-R and GF colonocytes. (B–D) Measurement of intermediary metabolites. Levels of acetyl-CoA (B), malate (C), and oxaloacetate (D) in CONV-R and GF. (E, F) Levels of mitochondrial NADH and NAD+ in CONV-R (E) and GF (F) colonocytes. The different scales reflect different amounts of material loaded and detected by HPLC-LC-MS. (G) Oxidative metabolism indicated by MitoTracker Red CM-H2XRos (red fluorescence; top panel) and MitoTracker Green FM (green fluorescence; middle panel) in colonocytes from CONV-R, CONV-R with NaN3 (sodium azide, negative control), or GF. (H) Quantification of oxidative metabolism in different experimental groups. (I) Spectral counts from quantitative mass spectrometry of ATP synthase F1α and β. Results in A–D, G, and H are displayed as mean ± SE and significant differences are identified (*p <0.05, **p <0.01). For panels A–D, 4 GF and 4 CONV-R mice were used. In panels E–H, 3 mice were used per condition.
Figure 4. Microbial Regulation of Energy Metabolism and Autophagy
(A–C) Western blot analysis of phosho-AMPK and AMPK (A), phospho-p27 (B) and LC3-I and -II (C) with histone H3 or β-actin as loading controls in CONV-R and GF. (D) Transmission electron micrographs of CONV-R and GF colonic epithelium. Lysosomes (L) and autophagosomes (a) are indicated, and arrows show double membrane of autophagasome in inset. For A–C, western blots are representative of 4 experiments using 2 CONV-R and 2 GF mice with 2 technical replicates each.
Figure 5. Colonization and Butyrate Rescue Diminished Oxidative Metabolism
(A) Mitochondrial respiration indicated by MitoTracker Red CM-H2XRos (red fluorescence; top panel) and MitoTracker Green FM (green fluorescence; middle panel) in colonocytes from CONV-R, CONV-D, B. fibrisolvens, GF, GF with 10 mM butyrate, and GF with both 10 mM butyrate and 500 µM etomoxir. (B) Quantification of mitochondrial respiration in different experimental groups. (C–D) Western blot analysis of phospho-p27 (C) and LC3-I and -II (D) with and without 10 mM butyrate in CONV-R and GF with β-actin as loading controls. (E) Western blot analysis of LC3-I and –II of GF, GF with 10 mM butyrate, and GF with 10 mM butyrate and 500 µM etomoxir. For panels A and B, a total of 3 mice per condition were used, and results are displayed as mean ± SE, with significant differences indicated (**p <0.01). For panels C–E, western blots are representative of 4 experiments using 2 CONV-R and 2 GF mice with 2 technical replicates each.
Figure 6. Microbial Regulation of Colonocyte Metabolism
Schematic showing that dietary fiber is fermented by microbes into butyrate in the lumen of the colon, which is transported into the colonocyte. In the colonocyte, butyrate promotes oxidative metabolism and inhibits autophagy. Based on transcriptome and proteome experiments, enzymes regulated by microbes are boxed. In all cases, boxed enzymes that function in β-oxidation and the TCA cycle are downregulated in GF colonocytes, revealing that microbes positively regulate their expression. Diminished ATP results in phosphorylation of AMPK and p27, which culminates in autophagy.
Comment in
- The microbial-mammalian metabolic axis: beyond simple metabolism.
Dumas ME. Dumas ME. Cell Metab. 2011 May 4;13(5):489-90. doi: 10.1016/j.cmet.2011.04.005. Cell Metab. 2011. PMID: 21531329
Similar articles
- Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes.
Donohoe DR, Wali A, Brylawski BP, Bultman SJ. Donohoe DR, et al. PLoS One. 2012;7(9):e46589. doi: 10.1371/journal.pone.0046589. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029553 Free PMC article. - Effect of germfree state on the capacities of isolated rat colonocytes to metabolize n-butyrate, glucose, and glutamine.
Cherbuy C, Darcy-Vrillon B, Morel MT, Pégorier JP, Duée PH. Cherbuy C, et al. Gastroenterology. 1995 Dec;109(6):1890-9. doi: 10.1016/0016-5085(95)90756-4. Gastroenterology. 1995. PMID: 7498654 - The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis.
Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. Liang J, et al. Nat Cell Biol. 2007 Feb;9(2):218-24. doi: 10.1038/ncb1537. Epub 2007 Jan 21. Nat Cell Biol. 2007. PMID: 17237771 - AMPK: guardian of metabolism and mitochondrial homeostasis.
Herzig S, Shaw RJ. Herzig S, et al. Nat Rev Mol Cell Biol. 2018 Feb;19(2):121-135. doi: 10.1038/nrm.2017.95. Epub 2017 Oct 4. Nat Rev Mol Cell Biol. 2018. PMID: 28974774 Free PMC article. Review. - Role of AMP-activated protein kinase in cancer therapy.
Rehman G, Shehzad A, Khan AL, Hamayun M. Rehman G, et al. Arch Pharm (Weinheim). 2014 Jul;347(7):457-68. doi: 10.1002/ardp.201300402. Epub 2014 Mar 28. Arch Pharm (Weinheim). 2014. PMID: 24677093 Review.
Cited by
- Butyrate: a bridge between intestinal flora and rheumatoid arthritis.
Cao Y, Chen J, Xiao J, Hong Y, Xu K, Zhu Y. Cao Y, et al. Front Immunol. 2024 Oct 16;15:1475529. doi: 10.3389/fimmu.2024.1475529. eCollection 2024. Front Immunol. 2024. PMID: 39478858 Free PMC article. Review. - Autophagy Improves Inflammatory Response in Sepsis Accompanied by Changes in Gut Microbiota.
Wang L, Wang W, Jiang G, Ke Z, Luo R, Tian W. Wang L, et al. Mediators Inflamm. 2024 Oct 18;2024:9550301. doi: 10.1155/2024/9550301. eCollection 2024. Mediators Inflamm. 2024. PMID: 39465181 Free PMC article. - Low-Molecular-Weight Compounds Produced by the Intestinal Microbiota and Cardiovascular Disease.
Cuervo L, McAlpine PL, Olano C, Fernández J, Lombó F. Cuervo L, et al. Int J Mol Sci. 2024 Sep 27;25(19):10397. doi: 10.3390/ijms251910397. Int J Mol Sci. 2024. PMID: 39408727 Free PMC article. Review. - Dietary and metabolic effects on intestinal stem cells in health and disease.
Shay JES, Yilmaz ÖH. Shay JES, et al. Nat Rev Gastroenterol Hepatol. 2024 Oct 2. doi: 10.1038/s41575-024-00980-7. Online ahead of print. Nat Rev Gastroenterol Hepatol. 2024. PMID: 39358589 Review. - The Impact of Yoyo Dieting and Resistant Starch on Weight Loss and Gut Microbiome in C57Bl/6 Mice.
Phuong-Nguyen K, O'Hely M, Kowalski GM, McGee SL, Aston-Mourney K, Connor T, Mahmood MQ, Rivera LR. Phuong-Nguyen K, et al. Nutrients. 2024 Sep 17;16(18):3138. doi: 10.3390/nu16183138. Nutrients. 2024. PMID: 39339738 Free PMC article.
References
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. - PubMed
- Boffa LC, Vidali G, Mann RS, Allfrey VG. Suppression of histone deacetylation in vivo and in vitro by sodium butyrate. J Biol Chem. 1978;253:3364–3366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA125237/CA/NCI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- R01 CA125237/CA/NCI NIH HHS/United States
- P30 DK034987/DK/NIDDK NIH HHS/United States
- P30 DK 34987/DK/NIDDK NIH HHS/United States
- P40RRO118603/PHS HHS/United States
- R01 CA125237-04/CA/NCI NIH HHS/United States
- P30-ES10126/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous