BrainAligner: 3D registration atlases of Drosophila brains - PubMed (original) (raw)
BrainAligner: 3D registration atlases of Drosophila brains
Hanchuan Peng et al. Nat Methods. 2011 Jun.
Abstract
Analyzing Drosophila melanogaster neural expression patterns in thousands of three-dimensional image stacks of individual brains requires registering them into a canonical framework based on a fiducial reference of neuropil morphology. Given a target brain labeled with predefined landmarks, the BrainAligner program automatically finds the corresponding landmarks in a subject brain and maps it to the coordinate system of the target brain via a deformable warp. Using a neuropil marker (the antibody nc82) as a reference of the brain morphology and a target brain that is itself a statistical average of data for 295 brains, we achieved a registration accuracy of 2 μm on average, permitting assessment of stereotypy, potential connectivity and functional mapping of the adult fruit fly brain. We used BrainAligner to generate an image pattern atlas of 2954 registered brains containing 470 different expression patterns that cover all the major compartments of the fly brain.
Figures
Figure 1
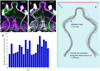
BrainAligner registers images of neurons from different brains onto a common coordinate system. (a–b) Maximum intensity projections of confocal images of a64-GAL4 and a74-GAL4 brains. Neurons are visualized by membrane-targeted GFP and brain morphology is visualized by staining with the antibody nc82. (c) Aligned and overlaid neuronal patterns of (a) and (b). (d) Alignment of many GAL4 expression patterns. Patterns of interest can be selected and displayed in the common coordinate system. R1 and R2, regions of interest. (e) V3D-AtlasViewer software for viewing the 3D pattern atlas. (f–h) Zoomed-in single-section views of R1 and R2 in (d). Scale bars in all sub-figures: 50 µm.
Figure 2
Schematic illustration of the BrainAligner algorithm. (a) BrainAligner performs a global alignment (G) followed by nonlinear local alignments (L) using landmarks. Scale bars: 50 µm. (b) The Reliable Landmark Matching (RLM) algorithm for detecting corresponding feature points in subject and target images. Dots of the same color indicate the matching landmarks; PT, a target brain landmark position; PS, a subject brain landmark; PMI, PINT, PCC, the best matching positions based on mutual information (MI), voxel intensity (INT), and correlation coefficient (CC) of local image patches. In the tetrahedron-pruning step, the landmarks in a subject image that clearly violate the relative position relationships of the target are discarded.
Figure 3
Stereotypy of neuronal morphology and reproducibility of GAL4 expression patterns. (a) Two aligned and overlaid examples (magenta and green) of the a278-GAL4 expression pattern, from different brains. Scale bar: 20 µm. (b) 3D reconstruction of the major neurite tracts in (a). Magenta and green, surface representations of the reconstructed tracts. Gray, GAL4 pattern. Scale bar: 20 µm. (c) 3D reconstructed neurite tracts (gray) from 20 aligned a278-GAL4 images, along with their mean tract model (red). (d) Average deviation of the mean tract model from each reconstructed tract.
Figure 4
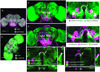
Expression pattern overlap by computational and biological methods. (a) Maximum intensity projection of a278-GAL4; UAS-mCD8-GFP. Scale bar: 100 µm. (b) Maximum intensity projection of LexAP036; lexop-CD2-GFP. Scale bar: 100 µm. (c) Aligned image of GAL4 and LexA expression patterns in (a) and (b), with a zoomed-in view to the right. Scale bar: 50 µm. (d) Co-expression of the GAL4 and LexA patterns, with a zoomed-in view to the right. Scale bar: 50 µm. Arrows indicate the 11 locations where colocalization of the two patterns was measured; the yellow arrow indicates a region of substantial overlap. (e–f) Cross-sectional views of single slices of the aligned (e) and co-expressed (f) samples at a position corresponding to the yellow arrow in (c) and (d). Scale bars: 25 µm.
Figure 5
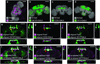
Comparison of computational alignment of separate brains with co-expression within the same brain. For all images, grey shows N-cadherin (N-Cad) labeling, which serves as the reference signal for alignment to the nc82-labeled target. Magenta, FasII antibody staining; green, GAL4 expression pattern (anti-GFP stain). (a) Wild-type w1118 adult brain. (b–d) Expression patterns of the indicated lines shown as maximum intensity projections of 20X confocal image stacks. (e, g, i) Cross-sectional views of computational alignments of FasII expression from (a) with GAL4 patterns from (b–d). (f, h, j) Matched cross-sectional views of brains expressing the GAL4 lines and labeled with both anti-GFP and anti-FasII to show biological co-localization. OK107 and 201Y expression patterns overlap with FasII (yellow arrows), but C232 expresses in adjacent but non-overlapping brain regions (red arrow). Scale bars, 100 µm.
Figure 6
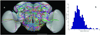
A 3D atlas of neurite tracts reconstructed from aligned GAL4 patterns. (a) 269 stereotyped neurite tracts and their distribution in the brain. The width of each tract equals the respective spatial deviation. The tracts are color-coded randomly for better visualization. Scale bar: 100 µm. (b) Distribution of the spatial deviation of the neurite tracts.
Comment in
- Bringing fly brains in line.
Huetteroth W, Waddell S. Huetteroth W, et al. Nat Methods. 2011 Jun;8(6):461-3. doi: 10.1038/nmeth.1615. Nat Methods. 2011. PMID: 21623350 No abstract available.
Similar articles
- Average shape standard atlas for the adult Drosophila ventral nerve cord.
Boerner J, Duch C. Boerner J, et al. J Comp Neurol. 2010 Jul 1;518(13):2437-55. doi: 10.1002/cne.22346. J Comp Neurol. 2010. PMID: 20503421 - Digital in vivo 3D atlas of the antennal lobe of Drosophila melanogaster.
Grabe V, Strutz A, Baschwitz A, Hansson BS, Sachse S. Grabe V, et al. J Comp Neurol. 2015 Feb 15;523(3):530-44. doi: 10.1002/cne.23697. Epub 2014 Nov 12. J Comp Neurol. 2015. PMID: 25327641 - An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy.
Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, Hartenstein V. Cardona A, et al. PLoS Biol. 2010 Oct 5;8(10):e1000502. doi: 10.1371/journal.pbio.1000502. PLoS Biol. 2010. PMID: 20957184 Free PMC article. - The insect central complex as model for heterochronic brain development-background, concepts, and tools.
Koniszewski ND, Kollmann M, Bigham M, Farnworth M, He B, Büscher M, Hütteroth W, Binzer M, Schachtner J, Bucher G. Koniszewski ND, et al. Dev Genes Evol. 2016 Jun;226(3):209-19. doi: 10.1007/s00427-016-0542-7. Epub 2016 Apr 7. Dev Genes Evol. 2016. PMID: 27056385 Free PMC article. Review. - deGradFP: A System to Knockdown GFP-Tagged Proteins.
Caussinus E, Affolter M. Caussinus E, et al. Methods Mol Biol. 2016;1478:177-187. doi: 10.1007/978-1-4939-6371-3_9. Methods Mol Biol. 2016. PMID: 27730581 Review.
Cited by
- LYNSU: automated 3D neuropil segmentation of fluorescent images for Drosophila brains.
Hsu KY, Shih CT, Chen NY, Lo CC. Hsu KY, et al. Front Neuroinform. 2024 Jul 29;18:1429670. doi: 10.3389/fninf.2024.1429670. eCollection 2024. Front Neuroinform. 2024. PMID: 39135968 Free PMC article. - FluoMALDI Microscopy: Matrix Co-Crystallization Simultaneously Enhances Fluorescence and MALDI Imaging.
Yang E, Shen XE, West-Foyle H, Hahm TH, Siegler MA, Brown DR, Johnson CC, Kim JH, Roker LA, Tressler CM, Barman I, Kuo SC, Glunde K. Yang E, et al. Adv Sci (Weinh). 2023 Dec;10(35):e2304343. doi: 10.1002/advs.202304343. Epub 2023 Oct 31. Adv Sci (Weinh). 2023. PMID: 37908150 Free PMC article. - Zebrafish brain atlases: a collective effort for a tiny vertebrate brain.
Légaré A, Lemieux M, Desrosiers P, De Koninck P. Légaré A, et al. Neurophotonics. 2023 Oct;10(4):044409. doi: 10.1117/1.NPh.10.4.044409. Epub 2023 Sep 30. Neurophotonics. 2023. PMID: 37786400 Free PMC article. - Full-Spectrum Neuronal Diversity and Stereotypy through Whole Brain Morphometry.
Liu Y, Jiang S, Li Y, Zhao S, Yun Z, Zhao ZH, Zhang L, Wang G, Chen X, Manubens-Gil L, Hang Y, Garcia-Forn M, Wang W, Rubeis S, Wu Z, Osten P, Gong H, Hawrylycz M, Mitra P, Dong H, Luo Q, Ascoli GA, Zeng H, Liu L, Peng H. Liu Y, et al. Res Sq [Preprint]. 2023 Jul 25:rs.3.rs-3146034. doi: 10.21203/rs.3.rs-3146034/v1. Res Sq. 2023. PMID: 37546984 Free PMC article. Preprint. - En bloc preparation of Drosophila brains enables high-throughput FIB-SEM connectomics.
Lu Z, Xu CS, Hayworth KJ, Pang S, Shinomiya K, Plaza SM, Scheffer LK, Rubin GM, Hess HF, Rivlin PK, Meinertzhagen IA. Lu Z, et al. Front Neural Circuits. 2022 Dec 16;16:917251. doi: 10.3389/fncir.2022.917251. eCollection 2022. Front Neural Circuits. 2022. PMID: 36589862 Free PMC article.
References
- Buchner E, Bader R, Buchner S, Cox J, Emson PC, Flory E, Heizmann CW, Hemm S, Hofbauer A, Oertel WH. Cell-specific immuno-probes for the brain of normal and mutant Drosophila melanogaster. I. Wildtype visual system. Cell and Tissue Research. 1988;253:357–370. - PubMed
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
- Luan H, White BH. Combinatorial methods for refined neuronal gene targeting. Current opinion in neurobiology. 2007;17:572–580. - PubMed
- Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and Inhibitory Switches for Courtship in the Brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases