Improved molecular replacement by density- and energy-guided protein structure optimization - PubMed (original) (raw)
. 2011 May 26;473(7348):540-3.
doi: 10.1038/nature09964. Epub 2011 May 1.
Thomas C Terwilliger, Randy J Read, Alexander Wlodawer, Gustav Oberdorfer, Ulrike Wagner, Eugene Valkov, Assaf Alon, Deborah Fass, Herbert L Axelrod, Debanu Das, Sergey M Vorobiev, Hideo Iwaï, P Raj Pokkuluri, David Baker
Affiliations
- PMID: 21532589
- PMCID: PMC3365536
- DOI: 10.1038/nature09964
Improved molecular replacement by density- and energy-guided protein structure optimization
Frank DiMaio et al. Nature. 2011.
Abstract
Molecular replacement procedures, which search for placements of a starting model within the crystallographic unit cell that best account for the measured diffraction amplitudes, followed by automatic chain tracing methods, have allowed the rapid solution of large numbers of protein crystal structures. Despite extensive work, molecular replacement or the subsequent rebuilding usually fail with more divergent starting models based on remote homologues with less than 30% sequence identity. Here we show that this limitation can be substantially reduced by combining algorithms for protein structure modelling with those developed for crystallographic structure determination. An approach integrating Rosetta structure modelling with Autobuild chain tracing yielded high-resolution structures for 8 of 13 X-ray diffraction data sets that could not be solved in the laboratories of expert crystallographers and that remained unsolved after application of an extensive array of alternative approaches. We estimate that the new method should allow rapid structure determination without experimental phase information for over half the cases where current methods fail, given diffraction data sets of better than 3.2 Å resolution, four or fewer copies in the asymmetric unit, and the availability of structures of homologous proteins with >20% sequence identity.
Figures
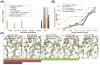
Figure 1
Examples of improvement in electron density and model quality. First row: Table 1 #6 (2.0 Å resolution); second row: Table 1 #7 (2.1 Å resolution); third row: Table 1 #12 (1.7 Å resolution). Left column: correct initial molecular replacement solution (not necessarily identifiable at this stage) using starting model and corresponding density. Middle column: energy optimized model and corresponding density. Right column: model and density following automatic building using the energy optimized model as the source of phase information. The final deposited structure is shown in yellow in each panel; the initial model, energy optimized model, and model after chain rebuilding are in red, green, and blue respectively. The sigma-A weighted 2mFo-DFc density contoured at 1.5σ is shown in gray.
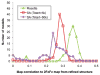
Figure 2
Method comparison. (A) Histogram of Rfree values after autobuilding for the seven difficult blind cases solved using the new approach (Table 1, set B). For most existing approaches, none of the cases yielded Rfree values under 50%; DEN was able to reduce Rfree to 45–49% for three of the structures. For all seven cases, Rosetta energy and density guided structure optimization led to Rfree values under 40%. (B) Dependence of success on sequence identity. The fraction of cases solved (Rfree after Autobuilding < 40%) is shown as a function of template sequence identity over the 18 blind cases and 59 benchmark cases. The new method is a clear improvement below 28% sequence identity. (C) Dependence of structure determination success on initial map quality. Sigma-A weighted 2mFo-DFc density maps (contoured at 1.5σ) computed from benchmark set templates with divergence from the native structure increasing from left to right are shown in grey; the solved crystal structure is shown in yellow. The correlation with the native density is shown above each panel. The solid green bar indicates structures the new approach was able to solve (Rfree<0.4); the red bar those that torsion-space refinement or DEN refinement is able to solve, and the purple bar those that can be solved directly using the template.
Figure 3
Comparison of the effectiveness of model diversification using Rosetta and simulated annealing. For blind case #6, 100 models were generated using either simulated annealing with a start temperature of 5000 K, simulated annealing with a start temperature of 50000 K, or Rosetta energy-and density-guided optimization. The correlation between 2mFo-DFc density maps computed from each structure and the final refined density was then computed; the starting model has a correlation of 0.29 and the distributions of the refined models are shown in the figure. Rosetta models have correlations better than the initial model much more often than simulated annealing.
Similar articles
- phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta.
Terwilliger TC, Dimaio F, Read RJ, Baker D, Bunkóczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. Terwilliger TC, et al. J Struct Funct Genomics. 2012 Jun;13(2):81-90. doi: 10.1007/s10969-012-9129-3. Epub 2012 Mar 15. J Struct Funct Genomics. 2012. PMID: 22418934 Free PMC article. - The JCSG MR pipeline: optimized alignments, multiple models and parallel searches.
Schwarzenbacher R, Godzik A, Jaroszewski L. Schwarzenbacher R, et al. Acta Crystallogr D Biol Crystallogr. 2008 Jan;64(Pt 1):133-40. doi: 10.1107/S0907444907050111. Epub 2007 Dec 5. Acta Crystallogr D Biol Crystallogr. 2008. PMID: 18094477 Free PMC article. - Improved technologies now routinely provide protein NMR structures useful for molecular replacement.
Mao B, Guan R, Montelione GT. Mao B, et al. Structure. 2011 Jun 8;19(6):757-66. doi: 10.1016/j.str.2011.04.005. Structure. 2011. PMID: 21645849 Free PMC article. - Molecular replacement: tricks and treats.
Abergel C. Abergel C. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2167-73. doi: 10.1107/S0907444913015291. Epub 2013 Oct 12. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189227 Free PMC article. Review. - Computational methods for remote homolog identification.
Wan XF, Xu D. Wan XF, et al. Curr Protein Pept Sci. 2005 Dec;6(6):527-46. doi: 10.2174/138920305774933231. Curr Protein Pept Sci. 2005. PMID: 16381602 Review.
Cited by
- Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems.
Adams PD, Baker D, Brunger AT, Das R, DiMaio F, Read RJ, Richardson DC, Richardson JS, Terwilliger TC. Adams PD, et al. Annu Rev Biophys. 2013;42:265-87. doi: 10.1146/annurev-biophys-083012-130253. Epub 2013 Feb 28. Annu Rev Biophys. 2013. PMID: 23451892 Free PMC article. Review. - Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM.
Fischer N, Neumann P, Konevega AL, Bock LV, Ficner R, Rodnina MV, Stark H. Fischer N, et al. Nature. 2015 Apr 23;520(7548):567-70. doi: 10.1038/nature14275. Epub 2015 Feb 23. Nature. 2015. PMID: 25707802 - Local error estimates dramatically improve the utility of homology models for solving crystal structures by molecular replacement.
Bunkóczi G, Wallner B, Read RJ. Bunkóczi G, et al. Structure. 2015 Feb 3;23(2):397-406. doi: 10.1016/j.str.2014.11.020. Epub 2015 Jan 22. Structure. 2015. PMID: 25619999 Free PMC article. - Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Lubkowski J, Chan W, Wlodawer A. Lubkowski J, et al. Sci Rep. 2019 Jul 30;9(1):11070. doi: 10.1038/s41598-019-46432-0. Sci Rep. 2019. PMID: 31363102 Free PMC article. - A brief history of macromolecular crystallography, illustrated by a family tree and its Nobel fruits.
Jaskolski M, Dauter Z, Wlodawer A. Jaskolski M, et al. FEBS J. 2014 Sep;281(18):3985-4009. doi: 10.1111/febs.12796. Epub 2014 Apr 17. FEBS J. 2014. PMID: 24698025 Free PMC article. Review.
References
- Rossmann MG. The Molecular Replacement Method. Gordon & Breach; New York: 1972.
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system (CNS): A new software system for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. - PubMed
- Vagin A, Teplyakov A. Molrep: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025.
Publication types
MeSH terms
Substances
Grants and funding
- 082961/WT_/Wellcome Trust/United Kingdom
- R01 GM092802/GM/NIGMS NIH HHS/United States
- U54 GM094586/GM/NIGMS NIH HHS/United States
- U54 GM074898/GM/NIGMS NIH HHS/United States
- U54 GM094597/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- GM074898/GM/NIGMS NIH HHS/United States
- U54GM074958/GM/NIGMS NIH HHS/United States
- P41RR002250/RR/NCRR NIH HHS/United States
- 5R01GM092802/GM/NIGMS NIH HHS/United States
- U54 GM074958/GM/NIGMS NIH HHS/United States
- P01 GM063210/GM/NIGMS NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources