Alternatively Spliced Genes as Biomarkers for Schizophrenia, Bipolar Disorder and Psychosis: A Blood-Based Spliceome-Profiling Exploratory Study - PubMed (original) (raw)
Alternatively Spliced Genes as Biomarkers for Schizophrenia, Bipolar Disorder and Psychosis: A Blood-Based Spliceome-Profiling Exploratory Study
S J Glatt et al. Curr Pharmacogenomics Person Med. 2009 Sep.
Abstract
OBJECTIVE: Transcriptomic biomarkers of psychiatric diseases obtained from a query of peripheral tissues that are clinically accessible (e.g., blood cells instead of post-mortem brain tissue) have substantial practical appeal to discern the molecular subtypes of common complex diseases such as major psychosis. To this end, spliceome-profiling is a new methodological approach that has considerable conceptual relevance for discovery and clinical translation of novel biomarkers for psychiatric illnesses. Advances in microarray technology now allow for improved sensitivity in measuring the transcriptome while simultaneously querying the "exome" (all exons) and "spliceome" (all alternatively spliced variants). The present study aimed to evaluate the feasibility of spliceome-profiling to discern transcriptomic biomarkers of psychosis. METHODS: We measured exome and spliceome expression in peripheral blood mononuclear cells from 13 schizophrenia patients, nine bipolar disorder patients, and eight healthy control subjects. Each diagnostic group was compared to each other, and the combined group of bipolar disorder and schizophrenia patients was also compared to the control group. Furthermore, we compared subjects with a history of psychosis to subjects without such history. RESULTS: After applying Bonferroni corrections for the 21,866 full-length gene transcripts analyzed, we found significant interactions between diagnostic group and exon identity, consistent with group differences in rates or types of alternative splicing. Relative to the control group, 18 genes in the bipolar disorder group, eight genes in the schizophrenia group, and 15 genes in the combined bipolar disorder and schizophrenia group appeared differentially spliced. Importantly, thirty-three genes showed differential splicing patterns between the bipolar disorder and schizophrenia groups. More frequent exon inclusion and/or over-expression was observed in psychosis. Finally, these observations are reconciled with an analysis of the ontologies, the pathways and the protein domains significantly over-represented among the alternatively spliced genes, several of which support prior discoveries. CONCLUSIONS: To our knowledge, this is the first blood-based spliceome-profiling study of schizophrenia and bipolar disorder to be reported. The battery of alternatively spliced genes and exons identified in this discovery-oriented exploratory study, if replicated, may have potential utility to discern the molecular subtypes of psychosis. Spliceome-profiling, as a new methodological approach in transcriptomics, warrants further work to evaluate its utility in personalized medicine. Potentially, this approach could also permit the future development of tissue-sampling methodologies in a form that is more acceptable to patients and thereby allow monitoring of dynamic and time-dependent plasticity in disease severity and response to therapeutic interventions in clinical psychiatry.
Conflict of interest statement
DUALITY/CONFLICT OF INTERESTS
None declared/applicable.
Figures
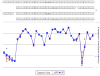
Fig. (1). Alternative Splicing of PTK2B: BPD vs. CNT
The figure illustrates a prototypical example of differential expression of alternatively spliced gene variants between two groups; in this case, BPD and CNT groups. The illustrated gene, PTK2B, produced the smallest _p_-value (8.19_e_−08) for the interaction of diagnostic group (BPD vs. CNT) and exon ID. The top panel shows the four known splice variants of the gene, while the bottom panel plots the raw microarray expression levels (signal intensities, unadjusted for covariates) of individual exons in the BPD and CNT groups (red triangles and blue squares, respectively). The lines representing signal intensity in each diagnostic group closely correspond to each other over most of the length of the gene, but intensities of some exons appeared to diverge, especially in the area of two known splicing sites. When controlling for all covariates, this divergence was statistically significant (*) at exons 2 (_p_=0.010), 31 (_p_=0.007), and 33(_p_=0.027), while the apparent decrease in expression of exon 29 (a known alternatively spliced exon) in the BPD group was not significant (_p_=0.299).
Similar articles
- NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences.
Atz ME, Rollins B, Vawter MP. Atz ME, et al. Psychiatr Genet. 2007 Apr;17(2):55-67. doi: 10.1097/YPG.0b013e328012d850. Psychiatr Genet. 2007. PMID: 17413444 Free PMC article. - Dysregulation of circRNA expression in the peripheral blood of individuals with schizophrenia and bipolar disorder.
Mahmoudi E, Green MJ, Cairns MJ. Mahmoudi E, et al. J Mol Med (Berl). 2021 Jul;99(7):981-991. doi: 10.1007/s00109-021-02070-6. Epub 2021 Mar 29. J Mol Med (Berl). 2021. PMID: 33782720 - Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia.
Cohen OS, Mccoy SY, Middleton FA, Bialosuknia S, Zhang-James Y, Liu L, Tsuang MT, Faraone SV, Glatt SJ. Cohen OS, et al. Schizophr Res. 2012 Dec;142(1-3):188-99. doi: 10.1016/j.schres.2012.09.015. Epub 2012 Oct 9. Schizophr Res. 2012. PMID: 23062752 Free PMC article. - The unstable trinucleotide repeat story of major psychosis.
Vincent JB, Paterson AD, Strong E, Petronis A, Kennedy JL. Vincent JB, et al. Am J Med Genet. 2000 Spring;97(1):77-97. doi: 10.1002/(sici)1096-8628(200021)97:1<77::aid-ajmg11>3.0.co;2-3. Am J Med Genet. 2000. PMID: 10813808 Review. - Specificity and Continuity of Schizophrenia and Bipolar Disorder: Relation to Biomarkers.
Yamada Y, Matsumoto M, Iijima K, Sumiyoshi T. Yamada Y, et al. Curr Pharm Des. 2020;26(2):191-200. doi: 10.2174/1381612825666191216153508. Curr Pharm Des. 2020. PMID: 31840595 Free PMC article. Review.
Cited by
- Acute prenatal exposure to a moderate dose of valproic acid increases social behavior and alters gene expression in rats.
Cohen OS, Varlinskaya EI, Wilson CA, Glatt SJ, Mooney SM. Cohen OS, et al. Int J Dev Neurosci. 2013 Dec;31(8):740-50. doi: 10.1016/j.ijdevneu.2013.09.002. Epub 2013 Sep 19. Int J Dev Neurosci. 2013. PMID: 24055786 Free PMC article. - Construction and analysis of the protein-protein interaction networks for schizophrenia, bipolar disorder, and major depression.
Lee SA, Tsao TT, Yang KC, Lin H, Kuo YL, Hsu CH, Lee WK, Huang KC, Kao CY. Lee SA, et al. BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S20. doi: 10.1186/1471-2105-12-S13-S20. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22373040 Free PMC article. - Blood-based gene-expression biomarkers of post-traumatic stress disorder among deployed marines: A pilot study.
Tylee DS, Chandler SD, Nievergelt CM, Liu X, Pazol J, Woelk CH, Lohr JB, Kremen WS, Baker DG, Glatt SJ, Tsuang MT; Marine Resiliency Study Investigators. Tylee DS, et al. Psychoneuroendocrinology. 2015 Jan;51:472-94. doi: 10.1016/j.psyneuen.2014.09.024. Epub 2014 Sep 30. Psychoneuroendocrinology. 2015. PMID: 25311155 Free PMC article. - Novel integrative genomic tool for interrogating lithium response in bipolar disorder.
Hunsberger JG, Chibane FL, Elkahloun AG, Henderson R, Singh R, Lawson J, Cruceanu C, Nagarajan V, Turecki G, Squassina A, Medeiros CD, Del Zompo M, Rouleau GA, Alda M, Chuang DM. Hunsberger JG, et al. Transl Psychiatry. 2015 Feb 3;5(2):e504. doi: 10.1038/tp.2014.139. Transl Psychiatry. 2015. PMID: 25646593 Free PMC article. - Using 3D epigenomic maps of primary olfactory neuronal cells from living individuals to understand gene regulation.
Rhie SK, Schreiner S, Witt H, Armoskus C, Lay FD, Camarena A, Spitsyna VN, Guo Y, Berman BP, Evgrafov OV, Knowles JA, Farnham PJ. Rhie SK, et al. Sci Adv. 2018 Dec 13;4(12):eaav8550. doi: 10.1126/sciadv.aav8550. eCollection 2018 Dec. Sci Adv. 2018. PMID: 30555922 Free PMC article.
References
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–92. - PubMed
- Cardno AG, Marshall EJ, Coid B, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry. 1999;56(2):162–8. - PubMed
- Kato T. Molecular genetics of bipolar disorder and depression. Psychiatry Clin Neurosci. 2007;61(1):3–19. - PubMed
Grants and funding
- R01 DA012846/DA/NIDA NIH HHS/United States
- R01 AG022982/AG/NIA NIH HHS/United States
- L30 DA022330-04/DA/NIDA NIH HHS/United States
- R25 MH081482/MH/NIMH NIH HHS/United States
- R01 MH071912/MH/NIMH NIH HHS/United States
- P50 MH081755-02/MH/NIMH NIH HHS/United States
- R01 MH065562/MH/NIMH NIH HHS/United States
- P30 MH062512/MH/NIMH NIH HHS/United States
- P50 MH081755/MH/NIMH NIH HHS/United States
- L30 DA022330/DA/NIDA NIH HHS/United States
- R01 AG022381/AG/NIA NIH HHS/United States
- R21 MH075027-02/MH/NIMH NIH HHS/United States
- R01 AG018386/AG/NIA NIH HHS/United States
- R01 MH079881/MH/NIMH NIH HHS/United States
- R25 MH074508/MH/NIMH NIH HHS/United States
- R41 MH079728/MH/NIMH NIH HHS/United States
- R21 MH075027/MH/NIMH NIH HHS/United States
- R01 MH085521-02/MH/NIMH NIH HHS/United States
- R01 MH085521/MH/NIMH NIH HHS/United States
- R01 DA018662/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources