Discovering biological progression underlying microarray samples - PubMed (original) (raw)
Discovering biological progression underlying microarray samples
Peng Qiu et al. PLoS Comput Biol. 2011 Apr.
Abstract
In biological systems that undergo processes such as differentiation, a clear concept of progression exists. We present a novel computational approach, called Sample Progression Discovery (SPD), to discover patterns of biological progression underlying microarray gene expression data. SPD assumes that individual samples of a microarray dataset are related by an unknown biological process (i.e., differentiation, development, cell cycle, disease progression), and that each sample represents one unknown point along the progression of that process. SPD aims to organize the samples in a manner that reveals the underlying progression and to simultaneously identify subsets of genes that are responsible for that progression. We demonstrate the performance of SPD on a variety of microarray datasets that were generated by sampling a biological process at different points along its progression, without providing SPD any information of the underlying process. When applied to a cell cycle time series microarray dataset, SPD was not provided any prior knowledge of samples' time order or of which genes are cell-cycle regulated, yet SPD recovered the correct time order and identified many genes that have been associated with the cell cycle. When applied to B-cell differentiation data, SPD recovered the correct order of stages of normal B-cell differentiation and the linkage between preB-ALL tumor cells with their cell origin preB. When applied to mouse embryonic stem cell differentiation data, SPD uncovered a landscape of ESC differentiation into various lineages and genes that represent both generic and lineage specific processes. When applied to a prostate cancer microarray dataset, SPD identified gene modules that reflect a progression consistent with disease stages. SPD may be best viewed as a novel tool for synthesizing biological hypotheses because it provides a likely biological progression underlying a microarray dataset and, perhaps more importantly, the candidate genes that regulate that progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
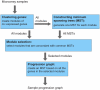
Figure 1. Sample Progression Discovery (SPD) framework.
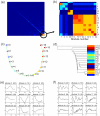
Figure 2. SPD applied to a cell cycle gene expression dataset.
(a) Based on an expression matrix of 3196 genes for 17 unordered samples from one cell cycle, SPD derived 154 modules and a progression similarity matrix between them. (b) Zoomed-in view of the progression similarity matrix highlights nine modules that are similar in terms of progression. (c) SPD constructed an overall MST to describe the common progression supported by the nine modules, showing a near perfect reconstruction of the correct time order. (d) Hierarchical clustering analysis did not recover the correct time order. In (c) and (d), Samples were color-coded according to the time points ( , hours), when the samples were taken. Blue corresponds to earlier time points; red corresponds to later time points. (e) The average expressions of the nine modules across the 17 time points show that, some of the nine modules were uncorrelated, i.e., modules 10 and 30, but SPD identified them as similar in terms of progression. (f) The mean expressions of the nine modules across all three cell cycles. The number in the parentheses above each plot is the number of genes in the corresponding module.
, hours), when the samples were taken. Blue corresponds to earlier time points; red corresponds to later time points. (e) The average expressions of the nine modules across the 17 time points show that, some of the nine modules were uncorrelated, i.e., modules 10 and 30, but SPD identified them as similar in terms of progression. (f) The mean expressions of the nine modules across all three cell cycles. The number in the parentheses above each plot is the number of genes in the corresponding module.
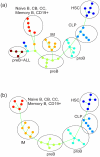
Figure 3. SPD applied to gene expression data of B-cell differentiation.
(a) Analysis based on all samples in this dataset. (b) Analysis of normal samples, with cancer samples and outliers next to cancer samples removed. Samples are color-coded according to their classification: HSC (violet), CLP (blue), proB (light blue), preB (green), immature (yellow), naiveB/CB/CC/memoryB/CD19+ (red), preB-ALL (brown). Circles are added to highlight each class of samples.
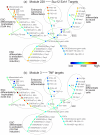
Figure 4. SPD applied to mouse embryonic stem cell differentiation data.
SPD revealed a landscape of mouse embryonic stem cell differentiation, where samples were perfectly ordered in time, with progressively later stages of differentiating cells radiating outwards from a core cluster of ESC samples. Circles were added to highlight each lineage. Nodes were color-coded by the expression level of a gene module (blue means low expression; green/yellow means medium; red means high expression). Module 228 was progressively induced in all differentiating lineages, and was enriched for Suz12 and Ezh1 targets. Module 3, enriched by TNF targets, was highly specifically regulated along one lineage, the trophoblast.
Figure 5. SPD applied to a prostate cancer dataset and derived a tree structure that describes the underlying progression.
(a) Nodes were color-coded according to their classification: normal, normal adjacent to tumor (NAP), tumor, metastatic samples (Mets). In (b), (c) and (d), nodes were color-coded using the average expression of modules 2, 32 and 19, respectively, in order to show how the expression of these modules gradually change during the progression.
Similar articles
- Discovering monotonic stemness marker genes from time-series stem cell microarray data.
Wang HW, Sun HJ, Chang TY, Lo HH, Cheng WC, Tseng GC, Lin CT, Chang SJ, Pal N, Chung IF. Wang HW, et al. BMC Genomics. 2015;16 Suppl 2(Suppl 2):S2. doi: 10.1186/1471-2164-16-S2-S2. Epub 2015 Jan 21. BMC Genomics. 2015. PMID: 25708300 Free PMC article. - Biclustering of microarray data with MOSPO based on crowding distance.
Liu J, Li Z, Hu X, Chen Y. Liu J, et al. BMC Bioinformatics. 2009 Apr 29;10 Suppl 4(Suppl 4):S9. doi: 10.1186/1471-2105-10-S4-S9. BMC Bioinformatics. 2009. PMID: 19426457 Free PMC article. - Dynamic biclustering of microarray data by multi-objective immune optimization.
Liu J, Li Z, Hu X, Chen Y, Park EK. Liu J, et al. BMC Genomics. 2011;12 Suppl 2(Suppl 2):S11. doi: 10.1186/1471-2164-12-S2-S11. Epub 2011 Jul 27. BMC Genomics. 2011. PMID: 21989068 Free PMC article. - Microarrays--identifying molecular portraits for prostate tumors with different Gleason patterns.
Mendes A, Scott RJ, Moscato P. Mendes A, et al. Methods Mol Med. 2008;141:131-51. doi: 10.1007/978-1-60327-148-6_8. Methods Mol Med. 2008. PMID: 18453088 Review. - Matrix factorisation methods applied in microarray data analysis.
Kossenkov AV, Ochs MF. Kossenkov AV, et al. Int J Data Min Bioinform. 2010;4(1):72-90. doi: 10.1504/ijdmb.2010.030968. Int J Data Min Bioinform. 2010. PMID: 20376923 Free PMC article. Review.
Cited by
- The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities.
Ng JW, Barrett LM, Wong A, Kuh D, Smith GD, Relton CL. Ng JW, et al. Genome Biol. 2012 Jun 29;13(6):246. doi: 10.1186/gb-2012-13-6-246. Genome Biol. 2012. PMID: 22747597 Free PMC article. - BitPhylogeny: a probabilistic framework for reconstructing intra-tumor phylogenies.
Yuan K, Sakoparnig T, Markowetz F, Beerenwinkel N. Yuan K, et al. Genome Biol. 2015 Feb 13;16(1):36. doi: 10.1186/s13059-015-0592-6. Genome Biol. 2015. PMID: 25786108 Free PMC article. - Inferring Multidimensional Rates of Aging from Cross-Sectional Data.
Pierson E, Koh PW, Hashimoto T, Koller D, Leskovec J, Eriksson N, Liang P. Pierson E, et al. Proc Mach Learn Res. 2019 Apr;89:97-107. Proc Mach Learn Res. 2019. PMID: 31538144 Free PMC article. - Design of a multi-signature ensemble classifier predicting neuroblastoma patients' outcome.
Cornero A, Acquaviva M, Fardin P, Versteeg R, Schramm A, Eva A, Bosco MC, Blengio F, Barzaghi S, Varesio L. Cornero A, et al. BMC Bioinformatics. 2012 Mar 28;13 Suppl 4(Suppl 4):S13. doi: 10.1186/1471-2105-13-S4-S13. BMC Bioinformatics. 2012. PMID: 22536959 Free PMC article. - Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers.
Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma'ayan A. Chen EY, et al. Bioinformatics. 2012 Jan 1;28(1):105-11. doi: 10.1093/bioinformatics/btr625. Epub 2011 Nov 10. Bioinformatics. 2012. PMID: 22080467 Free PMC article.
References
- Mandel E, Grosschedl R. Transcription control of early b cell differentiation. Curr Opin Immunol. 2010;22:161–167. - PubMed
- Filkov V, Skiena S, Zhi J. Analysis techniques for microarray time-series data. J Comput Biol. 2002;9:317–330. - PubMed
- Sacchi L, Larizza C, Magni P, Bellazzi R. Precedence temporal networks to represent temporal relationships in gene expression data. J Biomed Inform. 2007;40:761–774. - PubMed
- Zhu D, Hero A, Cheng H, Khanna R, Swaroop A. Network constrained clustering for gene microarray data. Bioinformatics. 2005;21:4014–4020. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases