Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes - PubMed (original) (raw)
Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes
Can Cenik et al. PLoS Genet. 2011 Apr.
Abstract
In higher eukaryotes, messenger RNAs (mRNAs) are exported from the nucleus to the cytoplasm via factors deposited near the 5' end of the transcript during splicing. The signal sequence coding region (SSCR) can support an alternative mRNA export (ALREX) pathway that does not require splicing. However, most SSCR-containing genes also have introns, so the interplay between these export mechanisms remains unclear. Here we support a model in which the furthest upstream element in a given transcript, be it an intron or an ALREX-promoting SSCR, dictates the mRNA export pathway used. We also experimentally demonstrate that nuclear-encoded mitochondrial genes can use the ALREX pathway. Thus, ALREX can also be supported by nucleotide signals within mitochondrial-targeting sequence coding regions (MSCRs). Finally, we identified and experimentally verified novel motifs associated with the ALREX pathway that are shared by both SSCRs and MSCRs. Our results show strong correlation between 5' untranslated region (5'UTR) intron presence/absence and sequence features at the beginning of the coding region. They also suggest that genes encoding secretory and mitochondrial proteins share a common regulatory mechanism at the level of mRNA export.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
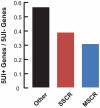
Figure 1. 5′UTR introns (5UIs) are depleted in genes that contain SSCRs and MSCRs.
Fraction of genes with 5UIs was plotted for SSCR, MSCR or other human protein coding genes.
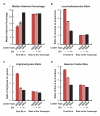
Figure 2. Adenine depletion in SSCRs is attenuated by 5UIs.
All bars represent the average from the first 69 nucleotides of all SSCRs-containing open reading frames from the human genome which either lacked (“−“) or had (“+”) 5UIs. As controls, these sequences were compared to the rest of the open reading frame, or analogous regions from open reading frames that lacked SSCRs (“other”). Error bars correspond to standard errors of the median or mean, as appropriate. (A) Median adenine percentage. Standard error of the median was determined by bootstrap-resampling. (B) The mean ratio between the number of encoded leucines to isoleucines. (C) The mean ratio between the number of encoded arginines to lysines. (D) The ratio of adenine- lacking, to adenine-containing codons for all amino acids that have both types of codons (L, V, A, P, S, G, and R). See Figure S3 for examples of adenine codon bias for specific amino acids (leucine and serine).
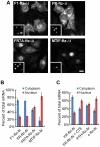
Figure 3. SSCRs derived from 5UI− genes promote mRNA nuclear export.
Capped and polyadenylated transcripts encoding various versions of ftz (see for all modified ftz sequences) were microinjected along with fluorescent 70 kD dextran into the nuclei of NIH 3T3 fibroblasts. After 1 hour, samples were fixed and mRNA was detected by FISH against ftz. (A) Representative micrographs of the mRNA distribution and fluorescent 70 kD dextran (inserts). Scale Bar = 15 µm. (B) Quantification of the cytoplasmic over the total fluorescence signal. Each bar represents an average of three experiments, each of which consisted of 15–30 cells. Error bars represent the standard deviation between the three experiments.
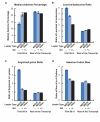
Figure 4. Adenine depletion in MSCRs derived from 5UI− genes.
As in Figure 2A, all bars represent the average from the first 69 nucleotides of all MSCRs-containing open reading frames from the human genome which either lacked (“–“) or had (“+”) 5UIs. As controls these sequences were compared to the rest of the open reading frame, or analogous regions from open reading frames that lacked SSCRs (“other”). Error bars correspond to the standard error of the median or mean, as appropriate. Panels A-D are as described for Figure 2. See Figure S3 for examples of adenine codon bias for specific amino acids (leucine and serine).
Figure 5. MSCRs derived from 5UI− genes promote mRNA nuclear export.
Transcripts encoding various versions of ftz (see Figure S4 for all modified ftz sequences) were microinjected along with fluorescent 70 kD dextran as in Figure 3. Panels A and B are as described for Figure 3. (C) DNA plasmids containing the indicated ftz genes and fluorescent 70 kD dextran were microinjected alone, or with the CTE viral RNA, into the nuclei of NIH 3T3 fibroblasts. Cells were allowed to express the ftz RNAs for 20 min then further transcription was inhibited by α-amanitin treatment. After allowing the mRNA to be exported for 2 hours, the cells were fixed and ftz mRNA was detected by fluorescence in situ hybridization. The distribution of ftz mRNA was quantified as described in Figure 3B.
Figure 6. Sequence features associated with ALREX elements.
(A) The ratio of thymine-lacking to thymine-containing codons for all amino acids that have both types (A, T, P, H, N, D, R, S, and G) was plotted for the first 69 nucleotides or the rest of the open reading frame of SSCR, MSCR or other genes. As in Figure 2A, all bars represent the average from open reading frames from the human genome which either lacked (“–”) or had (“+”) 5UIs. Error bars represent the standard error of the mean. (B) The position specific scoring matrix corresponding to the 6 nt motif was visualized using WebLogo (C) For each occurrence of the motif, the frame of translation is determined. The fraction of motif occurrences in all three possible frames were plotted for both 5UI− and 5UI+ SSCR-containing genes. (D) The distribution of the number of motifs in the set of SSCR-containing genes with 5UIs (negative set) and without 5UIs (positive set) were plotted. (E) For a given number of motif occurrences, the fraction of sequences in the positive versus negative set was plotted. Even though there were ∼2.5 times more sequences in the negative set, the fraction of sequences in the positive set with one or more occurrences of the motif was much higher compared to the fraction in the negative set. (F) The cumulative distribution of the motif occurrences were plotted for both sets (blue line for 5UI− genes and red line for 5UI+ genes) and for the uniform distribution (grey line). While the negative set did not differ from uniform distribution, the positive set displayed a left shift towards the 5′ of the transcript. (G) An ROC curve was generated to evaluate the discovered motif's predictive power in identifying the absence of 5UIs among MSCR-containing genes (see Materials and Methods). The performance of the CGSSGC motif is shown with the solid blue line, while the pale pink lines depict the performance of 50 randomly generated motifs. The boxplots represent the interquartile range of TPRs at a specified FPR for all 100,000 random motifs, and whiskers are drawn to 1.5 times the interquartile range. Outliers are not shown, and black horizontal line in each boxplot corresponds to the median TPR at the given FPR. The solid red line is the median performance of all 100,000 random motifs.
Figure 7. ALREX–associated motifs promote nuclear export of mRNA.
Various RNA motifs were inserted just after the start codon of the c-ftz-Δi construct. The nucleotide sequences of each RNA insert are depicted in (A). Note that the 4 silent adenine mutations used to convert M3 into 4A-M3 are indicated in bold. (B) DNA plasmids containing various versions of ftz were microinjected along with fluorescent 70 kD dextran into the nuclei of NIH 3T3 fibroblasts. After 20min, α-amanitin was added to halt further transcription and the cells were allowed to export the RNA for the indicated time periods. Cells were fixed, stained for ftz mRNA and export was quantified as in Figure 3B. Each bar represents an average of three separate experiments, each of which consisted of 15–30 cells. Error bars represent the standard deviation of the three experiments. (C) Representative cells from (B) microinjected with DNA plasmids containing either M3-ftz-Δi or 4A-M3-ftz-Δi and fixed 2 hours after α-amanitin treatment. Cells were imaged for ftz mRNA by FISH and fluorescent 70 kD dextran (insets). Scale Bar = 15 µm. (D) COS-7 cells were transfected with plasmids containing various versions of ftz and allowed to express the mRNA overnight. The cells were then fixed and stained for ftz mRNA by FISH and the cytoplasmic over the total fluorescence signal was quantified as in Figure 3B. Each bar represents an average of five separate experiments, each of which consisted of 20–50 cells. Error bars represent the standard error of the mean between the experiments.
Figure 8. Model describing 5′UTR intron effects on nuclear mRNA export by genes with SSCR/MSCRs.
(A) In the absence of 5UIs, the SSCR/MSCR is transcribed prior to any splicing event and as a consequence the transcript is exported via the ALREX pathway. (B) The presence of the 5UI results in recruitment of the splicesome and associated accessory proteins before the SSCR/MSCR is transcribed and hence the transcript is exported by the splicing-dependent pathway. As this transcript is not exported by the ALREX pathway, there is no selection pressure for the SSCR/MSCR to maintain specific nucleotide features associated with ALREX.
Similar articles
- The signal sequence coding region promotes nuclear export of mRNA.
Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. Palazzo AF, et al. PLoS Biol. 2007 Dec;5(12):e322. doi: 10.1371/journal.pbio.0050322. PLoS Biol. 2007. PMID: 18052610 Free PMC article. - ALREX-elements and introns: two identity elements that promote mRNA nuclear export.
Palazzo AF, Mahadevan K, Tarnawsky SP. Palazzo AF, et al. Wiley Interdiscip Rev RNA. 2013 Sep-Oct;4(5):523-33. doi: 10.1002/wrna.1176. Epub 2013 May 28. Wiley Interdiscip Rev RNA. 2013. PMID: 23913896 Review. - Positional requirements for the stimulation of mRNA nuclear export by ALREX-promoting elements.
Tarnawsky SP, Palazzo AF. Tarnawsky SP, et al. Mol Biosyst. 2012 Oct;8(10):2527-30. doi: 10.1039/c2mb25016k. Mol Biosyst. 2012. PMID: 22373716 - Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles.
Akef A, Zhang H, Masuda S, Palazzo AF. Akef A, et al. Nucleus. 2013 Jul-Aug;4(4):326-40. doi: 10.4161/nucl.26052. Epub 2013 Aug 8. Nucleus. 2013. PMID: 23934081 Free PMC article. - Insights into the nuclear export of murine leukemia virus intron-containing RNA.
Pessel-Vivares L, Houzet L, Lainé S, Mougel M. Pessel-Vivares L, et al. RNA Biol. 2015;12(9):942-9. doi: 10.1080/15476286.2015.1065375. RNA Biol. 2015. PMID: 26158194 Free PMC article. Review.
Cited by
- The GC-content at the 5' ends of human protein-coding genes is undergoing mutational decay.
Qiu Y, Kang YM, Korfmann C, Pouyet F, Eckford A, Palazzo AF. Qiu Y, et al. Genome Biol. 2024 Aug 13;25(1):219. doi: 10.1186/s13059-024-03364-x. Genome Biol. 2024. PMID: 39138526 Free PMC article. - mRNA nuclear export: how mRNA identity features distinguish functional RNAs from junk transcripts.
Palazzo AF, Qiu Y, Kang YM. Palazzo AF, et al. RNA Biol. 2024 Jan;21(1):1-12. doi: 10.1080/15476286.2023.2293339. Epub 2023 Dec 13. RNA Biol. 2024. PMID: 38091265 Free PMC article. Review. - A Study of 41 Canine Orthologues of Human Genes Involved in Monogenic Obesity Reveals Marker in the ADCY3 for Body Weight in Labrador Retrievers.
Sypniewski M, Szydlowski M. Sypniewski M, et al. Vet Sci. 2023 Jun 8;10(6):390. doi: 10.3390/vetsci10060390. Vet Sci. 2023. PMID: 37368776 Free PMC article. - mRNA in the Context of Protein Replacement Therapy.
Vavilis T, Stamoula E, Ainatzoglou A, Sachinidis A, Lamprinou M, Dardalas I, Vizirianakis IS. Vavilis T, et al. Pharmaceutics. 2023 Jan 3;15(1):166. doi: 10.3390/pharmaceutics15010166. Pharmaceutics. 2023. PMID: 36678793 Free PMC article. Review. - LAFITE Reveals the Complexity of Transcript Isoforms in Subcellular Fractions.
Zhang J, Lin X, Chen Y, Li TH, Lee AC, Chow EY, Cho WC, Chan TF. Zhang J, et al. Adv Sci (Weinh). 2023 Jan;10(3):e2203480. doi: 10.1002/advs.202203480. Epub 2022 Dec 3. Adv Sci (Weinh). 2023. PMID: 36461702 Free PMC article.
References
- Cenik C, Derti A, Mellor J, Berriz G, Roth F. Genome-wide functional analysis of human 5′ untranslated region introns. Genome Biology. 2010;11:R29. doi: 10.1186/gb-2010-11-3-r29. - DOI - PMC - PubMed
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, et al. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/S0378-1119(01)00674-6. - DOI - PubMed
- Hong X, Scofield DG, Lynch M. Intron Size, Abundance, and Distribution within Untranslated Regions of Genes. Mol Biol Evol. 2006;23:2392–2404. doi: 10.1093/molbev/msl111. - DOI - PubMed
- Sträβer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. - DOI - PubMed
- Cheng H, Dufu K, Lee C, Hsu JL, Dias A, et al. Human mRNA Export Machinery Recruited to the 5′ End of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG001715/HG/NHGRI NIH HHS/United States
- R21 MH087394/MH/NIMH NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- NS035611/NS/NINDS NIH HHS/United States
- U01 HG001715/HG/NHGRI NIH HHS/United States
- HG003224/HG/NHGRI NIH HHS/United States
- U01 HL081341/HL/NHLBI NIH HHS/United States
- U41 HG001715/HG/NHGRI NIH HHS/United States
- HG004233/HG/NHGRI NIH HHS/United States
- MH087394/MH/NIMH NIH HHS/United States
- P01 NS035611/NS/NINDS NIH HHS/United States
- HL081341/HL/NHLBI NIH HHS/United States
- P50 HG004233/HG/NHGRI NIH HHS/United States
- R01 HG003224/HG/NHGRI NIH HHS/United States
- HG001715/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources