Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes - PubMed (original) (raw)
Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes
Janine M Lamonica et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Acetylation of histones triggers association with bromodomain-containing proteins that regulate diverse chromatin-related processes. Although acetylation of transcription factors has been appreciated for some time, the mechanistic consequences are less well understood. The hematopoietic transcription factor GATA1 is acetylated at conserved lysines that are required for its stable association with chromatin. We show that the BET family protein Brd3 binds via its first bromodomain (BD1) to GATA1 in an acetylation-dependent manner in vitro and in vivo. Mutation of a single residue in BD1 that is involved in acetyl-lysine binding abrogated recruitment of Brd3 by GATA1, demonstrating that acetylation of GATA1 is essential for Brd3 association with chromatin. Notably, Brd3 is recruited by GATA1 to both active and repressed target genes in a fashion seemingly independent of histone acetylation. Anti-Brd3 ChIP followed by massively parallel sequencing in GATA1-deficient erythroid precursor cells and those that are GATA1 replete revealed that GATA1 is a major determinant of Brd3 recruitment to genomic targets within chromatin. A pharmacologic compound that occupies the acetyl-lysine binding pockets of Brd3 bromodomains disrupts the Brd3-GATA1 interaction, diminishes the chromatin occupancy of both proteins, and inhibits erythroid maturation. Together these findings provide a mechanism for GATA1 acetylation and suggest that Brd3 "reads" acetyl marks on nuclear factors to promote their stable association with chromatin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
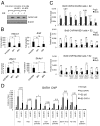
Fig. 1.
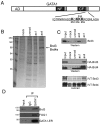
Brd3 is an acetyl-GATA1 associated protein. (A) Schematic of GATA1. AD, activation domain; gray boxes near the N-terminal (NF) and C-terminal (CF) zinc fingers represent conserved acetylated motifs. The sequence of the C-terminal acetylation motif is shown with four major acetylatable lysines in bold. (B) Colloidal blue stained gel of proteins bound to unmod, ac1, ac4 peptides, or to beads alone (mock). (C) Endogenous Brd3 (Top), stably expressed HA-Brd3 or HA-Brd4 (Middle), or IVT Brd3 and Brd4 bound to indicated (Bottom) GATA1 peptides were detected by Western blot or autoradiography. (D) Immunoprecipitation of GATA1-ER from G1E-ER4 cells followed by Western blot for GATA1, Brd3, and FOG1. Input equals 2.5%.
Fig. 2.
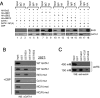
Acetylation-dependent interaction of BD1 with the C-terminal acetylation motif of GATA1. (A) Extracts from 293T cells transfected with indicated plasmids were immunoprecipitated with antibodies against HA or control IgG, followed by Western blotting with HA or GATA1 antibodies. Input equals 2.5%. (B) Immobilized GST-BD1, -BD2, and -Y72F were exposed to nuclear extracts from 293T cells expressing wild-type GATA1 or indicated GATA1 mutants with or without CBP, followed by anti-GATA1 Western blotting. Input equals 2.5%. (C) Immobilized GST-BD1, -BD2, and -Y72F were exposed to bulk chromatin from 293T cells, washed with 150 mM NaCl or 450 mM NaCl, followed by anti-acH4 Western blotting. Input equals 2.5%.
Fig. 3.
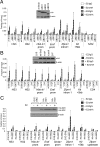
Cooccupancy of Brd3 and GATA1 at active and repressed target genes. (A) ChIP-qPCR with indicated antibodies or control (IgG) in G1E (−GATA1) and estradiol-treated G1E-ER4 (+GATA1) using primers specific for the β-globin locus. G1E-ER4 cells are definitive erythroid cells expressing adult (Hbb-b1) but not embryonic (Ey, bh1) globin genes. (B) ChIP-qPCR as in A using primers specific for the Kit locus. The data shown are the average of 3–6 experiments. Asterisks indicate p < 0.05. ns, not significant. Error bars represent standard deviation.
Fig. 4.
Association of Brd3 with GATA1 target sites requires acK binding by BD1. (A) Anti-HA ChIP of G1E-ER4 cells expressing HA-Brd3 or versions lacking BD1, BD2, or both bromodomains before or after estradiol (E2) treatment. (B) ChIP as in A in G1E-ER4 cells expressing HA-Brd3 with Y72F and/or Y347F substitutions. (C) BD1 is sufficient for recruitment by GATA1. HA-BD1 or HA-BD2 expressing G1E-ER4 cells were analyzed by anti-HA ChIP before and after treatment with E2. All constructs were expressed at comparable levels (Insets). Negative controls include 1 kb upstream of Hbb-b1 (IVR2), the 5′ transcribed region of CD4, or upstream of the Zfpm1 promoter. The data shown are the average of three to five experiments. All error bars represent standard deviation.
Fig. 5.
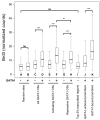
GATA1 occupancy is a major determinant for Brd3 occupancy genome-wide. The distributions of levels of Brd3 occupancy as determined by genome-wide ChIP in cells with or without active GATA1 are shown as box plots. Black bars denote the median number of normalized read counts, and boxes denote interquartile ranges (IQR). Whiskers demarcate the range of values excluding outliers as conventionally defined by > 1.5 × IQR. Statistical significance of difference was estimated by applying the Mann–Whitney U test on a random subset of data points (n = 400). “ns” indicates not significant; * indicates p < 0.05; ** indicates p < 10-5; *** indicates p < 10-10.
Fig. 6.
Effects of BD1 or BD2 on GATA1 function. (A) Control Western blot showing that expression of HA-BD1 or HA-BD2 does not affect GATA1-ER levels. (B) qRT-PCR measuring fold induction of indicated GATA1 target genes in G1E-ER4 cells expressing HA-BD1 or HA-BD2 before and after E2 treatment. Data are the average of five independent experiments. (C) Anti-Brd3 ChIP in parental G1E-ER4 cells (Top) or cells expressing HA-BD1 (Middle), HA-BD2 (Bottom). Data are the average of three independent experiments. (D) Anti-GATA1 ChIP comparing GATA1 occupancy in parental G1E-ER4 cells and HA-BD1 expressing cells. Data are the average of three to four independent experiments. Asterisks indicate p < 0.05. ns, not significant. All error bars denote standard deviation.
Fig. 7.
19X disrupts the interaction between Brd3 and acetylated GATA1, blocks erythroid maturation, and impairs GATA1 binding to chromatin. (A) GST-BD1 was exposed to nuclear extracts from 293T cells expressing GATA1 and CBP in the presence of the indicated concentrations of 19X. Bound material was analyzed by anti-GATA1 Western blot. Input equals 2.5%. (B) GATA1 peptides were incubated with nuclear extracts from 293T cells expressing full-length HA-Brd3 in the presence of the indicated concentrations of 19X. Retained material was probed by anti-HA Western blotting. Input equals 2.5%. (C) GATA1 peptide binding assays with GST-BD1 in the presence of varying concentrations of 19X. Bound material was analyzed by Western blotting using GST antibodies. Input equals 4%. (D) Anti-HA immunoprecipitation of nuclear extracts from 293T cells expressing GATA1 and HA-Brd3 in the presence of 19X, followed by Western blotting with anti-GATA1 or anti-HA antibodies. Input equals 2.5%. (E) qRT-PCR measuring fold changes in mRNA levels of GATA1 target genes Hbb-b1, Eraf, Slc4a1, and Zfpm1 (Right) and control genes Pabpc1 and Brd3 (Left) in G1E and estradiol-treated G1E-ER4 cells. Prior to harvesting, cells were grown in the presence of the indicated concentrations of 19X for 24 h. The data shown are the average of two to four independent experiments. (F) ChIP-qPCR analysis of G1E and estradiol-treated G1E-ER4 grown in the presence or absence of 1 μM 19X for 24 h. The data shown are the average of three to four independent experiments. Asterisks indicate p < 0.05. ns, not significant. All error bars denote standard deviation.
Fig. 1.
Acetylated GATA1 interacts with the bromodomain protein Brd3. The hematopoietic transcription factor GATA1 is acetylated on lysine clusters near its zinc finger domains (Left and Center). Acetylated GATA1 specifically binds to the first bromodomain of BET family member Brd3 (Right). The resulting Brd3-containing GATA1 complex is competent to stably bind to cognate WGATAR sites present in the regulatory regions of its target genes and regulate erythroid transcription.
Similar articles
- Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3.
Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Gamsjaeger R, et al. Mol Cell Biol. 2011 Jul;31(13):2632-40. doi: 10.1128/MCB.05413-11. Epub 2011 May 9. Mol Cell Biol. 2011. PMID: 21555453 Free PMC article. - Cooperative binding of two acetylation marks on a histone tail by a single bromodomain.
Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Müller CW, Petosa C. Morinière J, et al. Nature. 2009 Oct 1;461(7264):664-8. doi: 10.1038/nature08397. Nature. 2009. PMID: 19794495 - Functions of BET proteins in erythroid gene expression.
Stonestrom AJ, Hsu SC, Jahn KS, Huang P, Keller CA, Giardine BM, Kadauke S, Campbell AE, Evans P, Hardison RC, Blobel GA. Stonestrom AJ, et al. Blood. 2015 Apr 30;125(18):2825-34. doi: 10.1182/blood-2014-10-607309. Epub 2015 Feb 18. Blood. 2015. PMID: 25696920 Free PMC article. - The bromodomain: from epigenome reader to druggable target.
Sanchez R, Meslamani J, Zhou MM. Sanchez R, et al. Biochim Biophys Acta. 2014 Aug;1839(8):676-85. doi: 10.1016/j.bbagrm.2014.03.011. Epub 2014 Mar 28. Biochim Biophys Acta. 2014. PMID: 24686119 Free PMC article. Review. - Acetyllysine-binding and function of bromodomain-containing proteins in chromatin.
Dyson MH, Rose S, Mahadevan LC. Dyson MH, et al. Front Biosci. 2001 Aug 1;6:D853-65. doi: 10.2741/dyson. Front Biosci. 2001. PMID: 11487465 Review.
Cited by
- Effect of BET Missense Mutations on Bromodomain Function, Inhibitor Binding and Stability.
Lori L, Pasquo A, Lori C, Petrosino M, Chiaraluce R, Tallant C, Knapp S, Consalvi V. Lori L, et al. PLoS One. 2016 Jul 12;11(7):e0159180. doi: 10.1371/journal.pone.0159180. eCollection 2016. PLoS One. 2016. PMID: 27403962 Free PMC article. - Genomic Identification, Evolution, and Expression Analysis of Bromodomain Genes Family in Buffalo.
Zhang J, Huang L, Zhang P, Huang X, Yang W, Liu R, Sun Q, Lu Y, Zhang M, Fu Q. Zhang J, et al. Genes (Basel). 2022 Jan 1;13(1):103. doi: 10.3390/genes13010103. Genes (Basel). 2022. PMID: 35052443 Free PMC article. - Molecular insights into the recognition of N-terminal histone modifications by the BRPF1 bromodomain.
Poplawski A, Hu K, Lee W, Natesan S, Peng D, Carlson S, Shi X, Balaz S, Markley JL, Glass KC. Poplawski A, et al. J Mol Biol. 2014 Apr 17;426(8):1661-76. doi: 10.1016/j.jmb.2013.12.007. Epub 2013 Dec 12. J Mol Biol. 2014. PMID: 24333487 Free PMC article. - Interrogating Histone Acetylation and BRD4 as Mitotic Bookmarks of Transcription.
Behera V, Stonestrom AJ, Hamagami N, Hsiung CC, Keller CA, Giardine B, Sidoli S, Yuan ZF, Bhanu NV, Werner MT, Wang H, Garcia BA, Hardison RC, Blobel GA. Behera V, et al. Cell Rep. 2019 Apr 9;27(2):400-415.e5. doi: 10.1016/j.celrep.2019.03.057. Cell Rep. 2019. PMID: 30970245 Free PMC article. - Structural basis and specificity of acetylated transcription factor GATA1 recognition by BET family bromodomain protein Brd3.
Gamsjaeger R, Webb SR, Lamonica JM, Billin A, Blobel GA, Mackay JP. Gamsjaeger R, et al. Mol Cell Biol. 2011 Jul;31(13):2632-40. doi: 10.1128/MCB.05413-11. Epub 2011 May 9. Mol Cell Biol. 2011. PMID: 21555453 Free PMC article.
References
- Zeng L, Zhou MM. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. - PubMed
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 DK007780/DK/NIDDK NIH HHS/United States
- RC2 HG005573/HG/NHGRI NIH HHS/United States
- T32 DK07780/DK/NIDDK NIH HHS/United States
- R01 DK065806/DK/NIDDK NIH HHS/United States
- R01 DK054937/DK/NIDDK NIH HHS/United States
- T32 HL007971/HL/NHLBI NIH HHS/United States
- DK054937/DK/NIDDK NIH HHS/United States
- T32 HL007971-07/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases