Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1 - PubMed (original) (raw)
Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1
Balyn W Zaro et al. Proc Natl Acad Sci U S A. 2011.
Abstract
The dynamic modification of nuclear and cytoplasmic proteins by the monosaccharide N-acetyl-glucosamine (GlcNAc) continues to emerge as an important regulator of many biological processes. Herein we describe the development of an alkynyl-modified GlcNAc analog (GlcNAlk) as a new chemical reporter of O-GlcNAc modification in living cells. This strategy is based on metabolic incorporation of reactive functionality into the GlcNAc biosynthetic pathway. When combined with the Cu(I)-catalyzed [3 + 2] azide-alkyne cycloaddition, this chemical reporter allowed for the robust in-gel fluorescent visualization of O-GlcNAc and affinity enrichment and identification of O-GlcNAc-modified proteins. Using in-gel fluorescence detection, we characterized the metabolic fates of GlcNAlk and the previously reported azido analog, GlcNAz. We confirmed previous results that GlcNAz can be metabolically interconverted to GalNAz, whereas GlcNAlk does not, thereby yielding a more specific metabolic reporter of O-GlcNAc modification. We also used GlcNAlk, in combination with a biotin affinity tag, to identify 374 proteins, 279 of which were not previously reported, and we subsequently confirmed the enrichment of three previously uncharacterized proteins. Finally we confirmed the O-GlcNAc modification of the ubiquitin ligase NEDD4-1, the first reported glycosylation of this protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
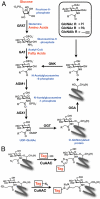
The HBP and chemical reporters of O-GlcNAc modification. (A) Glucose, glutamine, and acetyl-CoA are transformed by the metabolic enzymes of the HBP to UDP-GlcNAc, which can be used by OGT to modify protein substrates and reversed by OGA. Per-_O_-acetylated analogues (Ac4GlcNAz and Ac4GlcNAlk) are deacetylated by intracellular esterases, and then enter the GlcNAc salvage pathway. (B) GlcNAz and GlcNAlk bearing proteins can be covalently modified with detection tags using CuAAC.
Fig. 2.
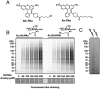
Fluorescent detection of O-GlcNAc-modified proteins. (A) Fluorescent-tags Alk-Rho and Az-Rho for CuAAC-dependent detection of GlcNAz and GlcNAlk (B) HEK293 cells were treated with the indicated concentrations of Ac4GlcNAz or Ac4GlcNAlk for 16 h and analyzed by in-gel fluorescence scanning. Coomassie blue staining demonstrated equal protein loading. (C) Higher contrast of lanes corresponding to 0 μM in B to allow for comparison of background levels.
Fig. 3.
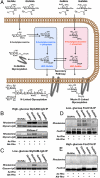
Characterizing GlcNAz and GlcNAlk protein labeling. (A) Monosaccharide chemical reports have several possible metabolic fates. GlcNAz and GlcNAlk can enter the glucosamine salvage pathway (blue pathway) and potentially label O-GlcNAc-modified proteins and N-linked glycans. They could also be reversibly converted to the corresponding GalNAc analogs, GalNAz and GalNAlk (red pathway), resulting in the potential labeling of mucin-type O-linked glycosylation. (B and C) COS-7 cells were transfected with a plasmid encoding GlyCAM-IgG, treated with the indicated chemical reporter, and analyzed by in-gel fluorescence scanning. PNGase-F treatment prior to CuAAC was performed to selectively remove N-linked glycans. (D and E) COS-7 cells were transfected with a plasmid encoding FoxO1A, treated with the indicated chemical reporter, and analyzed by in-gel fluorescence. For comparison experiments B and C, as well as experiments D and E, fluorescent levels were measured and normalized simultaneously.
Fig. 4.
Identification of O-GlcNAlk-modified proteins. (A) O-GlcNAlk-modified proteins were enriched from NIH3T3 cells treated with Ac4GlcNAlk (200 μM) using azido-azo-biotin and analyzed by Western blotting. (B) Known O-GlcNAcylated proteins p62 and α-B crystallin were immunoprecipitated from cells treated with Ac4GlcNAlk (200 μM) and analyzed by in-gel fluorescence. (C) NEDD4-1 was selectively enriched from cells treated with Ac4GlcNAlk (200 μM) and analyzed by in-gel fluorescence. (D) NEDD4-1 was immunoprecipitated from cells and analyzed by Western blotting.
Similar articles
- Comparative analysis of Cu (I)-catalyzed alkyne-azide cycloaddition (CuAAC) and strain-promoted alkyne-azide cycloaddition (SPAAC) in O-GlcNAc proteomics.
Li S, Zhu H, Wang J, Wang X, Li X, Ma C, Wen L, Yu B, Wang Y, Li J, Wang PG. Li S, et al. Electrophoresis. 2016 Jun;37(11):1431-6. doi: 10.1002/elps.201500491. Epub 2016 Mar 1. Electrophoresis. 2016. PMID: 26853435 Free PMC article. - [Comparison of the performance of secretome analysis based on metabolic labeling by three unnatural sugars].
Mao Y, Zheng J, Feng S, Tian R. Mao Y, et al. Se Pu. 2021 Oct;39(10):1086-1093. doi: 10.3724/SP.J.1123.2021.04017. Se Pu. 2021. PMID: 34505430 Free PMC article. Chinese. - Identification of new O-GlcNAc modified proteins using a click-chemistry-based tagging.
Gurcel C, Vercoutter-Edouart AS, Fonbonne C, Mortuaire M, Salvador A, Michalski JC, Lemoine J. Gurcel C, et al. Anal Bioanal Chem. 2008 Apr;390(8):2089-97. doi: 10.1007/s00216-008-1950-y. Epub 2008 Mar 28. Anal Bioanal Chem. 2008. PMID: 18369606 - Glycosylation of the nuclear pore.
Li B, Kohler JJ. Li B, et al. Traffic. 2014 Apr;15(4):347-61. doi: 10.1111/tra.12150. Epub 2014 Feb 13. Traffic. 2014. PMID: 24423194 Free PMC article. Review. - The Utilities of Chemical Reactions and Molecular Tools for O-GlcNAc Proteomic Studies.
Kim EJ. Kim EJ. Chembiochem. 2015 Jul 6;16(10):1397-409. doi: 10.1002/cbic.201500183. Epub 2015 Jun 11. Chembiochem. 2015. PMID: 26096757 Review.
Cited by
- Enzyme-mediated methodology for the site-specific radiolabeling of antibodies based on catalyst-free click chemistry.
Zeglis BM, Davis CB, Aggeler R, Kang HC, Chen A, Agnew BJ, Lewis JS. Zeglis BM, et al. Bioconjug Chem. 2013 Jun 19;24(6):1057-67. doi: 10.1021/bc400122c. Epub 2013 May 30. Bioconjug Chem. 2013. PMID: 23688208 Free PMC article. - Cell-type-specific labeling and profiling of glycans in living mice.
Fan X, Song Q, Sun DE, Hao Y, Wang J, Wang C, Chen X. Fan X, et al. Nat Chem Biol. 2022 Jun;18(6):625-633. doi: 10.1038/s41589-022-01016-4. Epub 2022 May 5. Nat Chem Biol. 2022. PMID: 35513511 - Recent development of analytical methods for disease-specific protein _O_-GlcNAcylation.
Hu W, Zhang G, Zhou Y, Xia J, Zhang P, Xiao W, Xue M, Lu Z, Yang S. Hu W, et al. RSC Adv. 2022 Dec 21;13(1):264-280. doi: 10.1039/d2ra07184c. eCollection 2022 Dec 19. RSC Adv. 2022. PMID: 36605671 Free PMC article. Review. - Mechanistic Investigation and Multiplexing of Liposome-Assisted Metabolic Glycan Labeling.
Sun Y, Hong S, Xie R, Huang R, Lei R, Cheng B, Sun DE, Du Y, Nycholat CM, Paulson JC, Chen X. Sun Y, et al. J Am Chem Soc. 2018 Mar 14;140(10):3592-3602. doi: 10.1021/jacs.7b10990. Epub 2018 Mar 2. J Am Chem Soc. 2018. PMID: 29446631 Free PMC article. - Deciphering the Functions of Protein O-GlcNAcylation with Chemistry.
Worth M, Li H, Jiang J. Worth M, et al. ACS Chem Biol. 2017 Feb 17;12(2):326-335. doi: 10.1021/acschembio.6b01065. Epub 2017 Jan 19. ACS Chem Biol. 2017. PMID: 28055183 Free PMC article. Review.
References
- Zachara NE, Hart GW. The emerging significance of O-GlcNAc in cellular regulation. Chem Rev. 2002;102:431–438. - PubMed
- Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005;312:re13. - PubMed
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources