Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes - PubMed (original) (raw)
Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes
Maria Anisimova et al. Syst Biol. 2011 Oct.
Abstract
Phylogenetic inference and evaluating support for inferred relationships is at the core of many studies testing evolutionary hypotheses. Despite the popularity of nonparametric bootstrap frequencies and Bayesian posterior probabilities, the interpretation of these measures of tree branch support remains a source of discussion. Furthermore, both methods are computationally expensive and become prohibitive for large data sets. Recent fast approximate likelihood-based measures of branch supports (approximate likelihood ratio test [aLRT] and Shimodaira-Hasegawa [SH]-aLRT) provide a compelling alternative to these slower conventional methods, offering not only speed advantages but also excellent levels of accuracy and power. Here we propose an additional method: a Bayesian-like transformation of aLRT (aBayes). Considering both probabilistic and frequentist frameworks, we compare the performance of the three fast likelihood-based methods with the standard bootstrap (SBS), the Bayesian approach, and the recently introduced rapid bootstrap. Our simulations and real data analyses show that with moderate model violations, all tests are sufficiently accurate, but aLRT and aBayes offer the highest statistical power and are very fast. With severe model violations aLRT, aBayes and Bayesian posteriors can produce elevated false-positive rates. With data sets for which such violation can be detected, we recommend using SH-aLRT, the nonparametric version of aLRT based on a procedure similar to the Shimodaira-Hasegawa tree selection. In general, the SBS seems to be excessively conservative and is much slower than our approximate likelihood-based methods.
Figures
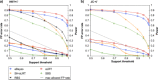
FIGURE 1.
FP error rate (continuous lines) and power (dotted lines) of branch support methods. Data are simulated with 100 taxa, 600 nucleotides under the covarion model and analyzed using incorrect models: (a) HKY + Γ and (b) JC + Γ.
FIGURE 2.
Probabilistic interpretation is rarely achieved. Inferred average support of a clade is plotted against the true probability under the true (HKY + Γ) and the incorrect (JC + Γ) models.
FIGURE 3.
Bayesian PP compared with aBayes supports, and their distributions in real data: (a) DNA data 1–8 from Table 1, analyzed assuming HKY + Γ; (b) AA data 9–16 from Table 1, analyzed assuming WAG + Γ.
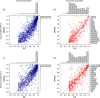
FIGURE 4.
Bayesian PP compared with aBayes supports and their distributions in simulations: (a) for correctly inferred branches under HKY + Γ; (b) for incorrectly inferred branches under HKY + Γ; (c) for correctly inferred branches under JC + Γ; (d) for incorrectly inferred branches under JC + Γ.
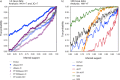
FIGURE 5.
Comparison of branch support measures on the nsf2-F gene: (a) Metazoan phylogeny reconstructed for the nsf2-F gene with ML using PHYML; (b) estimated branch supports corresponding to reconstructed branches, and (c) the hypothesized species tree (Guindon and Gascuel 2003; Lartillot et al. 2007).
Similar articles
- Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative.
Anisimova M, Gascuel O. Anisimova M, et al. Syst Biol. 2006 Aug;55(4):539-52. doi: 10.1080/10635150600755453. Syst Biol. 2006. PMID: 16785212 - Predicting Phylogenetic Bootstrap Values via Machine Learning.
Wiegert J, Höhler D, Haag J, Stamatakis A. Wiegert J, et al. Mol Biol Evol. 2024 Oct 4;41(10):msae215. doi: 10.1093/molbev/msae215. Mol Biol Evol. 2024. PMID: 39418337 Free PMC article. - Divergent maximum-likelihood-branch-support values for polytomies.
Simmons MP, Norton AP. Simmons MP, et al. Mol Phylogenet Evol. 2014 Apr;73:87-96. doi: 10.1016/j.ympev.2014.01.018. Epub 2014 Feb 4. Mol Phylogenet Evol. 2014. PMID: 24503481 - Statistical inference for stochastic simulation models--theory and application.
Hartig F, Calabrese JM, Reineking B, Wiegand T, Huth A. Hartig F, et al. Ecol Lett. 2011 Aug;14(8):816-27. doi: 10.1111/j.1461-0248.2011.01640.x. Epub 2011 Jun 17. Ecol Lett. 2011. PMID: 21679289 Review.
Cited by
- Pan-mitogenomics reveals the genetic basis of cytonuclear conflicts in citrus hybridization, domestication, and diversification.
Wang N, Li C, Kuang L, Wu X, Xie K, Zhu A, Xu Q, Larkin RM, Zhou Y, Deng X, Guo W. Wang N, et al. Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2206076119. doi: 10.1073/pnas.2206076119. Epub 2022 Oct 19. Proc Natl Acad Sci U S A. 2022. PMID: 36260744 Free PMC article. - Complete mitochondrial DNA sequences of the threadfin cichlid (Petrochromis trewavasae) and the blunthead cichlid (Tropheus moorii) and patterns of mitochondrial genome evolution in cichlid fishes.
Fischer C, Koblmüller S, Gülly C, Schlötterer C, Sturmbauer C, Thallinger GG. Fischer C, et al. PLoS One. 2013 Jun 24;8(6):e67048. doi: 10.1371/journal.pone.0067048. Print 2013. PLoS One. 2013. PMID: 23826193 Free PMC article. - Intra-Patient Evolution of HIV-2 Molecular Properties.
Palm AA, Esbjörnsson J, Kvist A, Månsson F, Biague A, Norrgren H, Jansson M, Medstrand P. Palm AA, et al. Viruses. 2022 Nov 4;14(11):2447. doi: 10.3390/v14112447. Viruses. 2022. PMID: 36366545 Free PMC article. - Using multiple sampling strategies to estimate SARS-CoV-2 epidemiological parameters from genomic sequencing data.
Inward RPD, Parag KV, Faria NR. Inward RPD, et al. Nat Commun. 2022 Sep 23;13(1):5587. doi: 10.1038/s41467-022-32812-0. Nat Commun. 2022. PMID: 36151084 Free PMC article. - Genomic remnants of ancestral methanogenesis and hydrogenotrophy in Archaea drive anaerobic carbon cycling.
Adam PS, Kolyfetis GE, Bornemann TLV, Vorgias CE, Probst AJ. Adam PS, et al. Sci Adv. 2022 Nov 4;8(44):eabm9651. doi: 10.1126/sciadv.abm9651. Epub 2022 Nov 4. Sci Adv. 2022. PMID: 36332026 Free PMC article.
References
- Akaike H. Information theory and an extension of the maximum likelihood principle. Second International Symposium on Information Theory. In: Petrov BN, Csaki F, editors. Budapest (Hungary) Akademiai Kiado; 1973. pp. 267–281.
- Aldous D. Probability distributions of cladograms. In: Aldous D, Pemantle R, editors. Random discrete structures. New York: Springer-Verlag; 1996. pp. 1–18.
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. - PubMed
- Baum DA, Smith SD, Donovan SS. Evolution. The tree-thinking challenge. Science. 2005;310:979–980. - PubMed
- Berry V, Gascuel O. On the interpretation of bootstrap trees: appropriate threshold of clade selection and induced gain. Mol. Biol. Evol. 1996;13:999–1011.