The cancerous translation apparatus - PubMed (original) (raw)
Review
The cancerous translation apparatus
Craig R Stumpf et al. Curr Opin Genet Dev. 2011 Aug.
Abstract
Deregulations in translational control are critical features of cancer initiation and progression. Activation of key oncogenic pathways promotes rapid and dramatic translational reprogramming, not simply by increasing overall protein synthesis, but also by modulating specific mRNA networks that promote cellular transformation. Additionally, ribosomopathies caused by mutations in ribosome components alter translational regulation leading to specific pathological features, including cancer susceptibility. Exciting advances in our understanding of translational control in cancer have illuminated a striking specificity innate to the translational apparatus. Characterizing this specificity will provide novel insights into how cells normally utilize translational control to modulate gene expression, how it is deregulated in cancer, and how these processes can be targeted to develop new cancer therapies.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
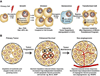
Figure 1. Deregulations in translational control can contribute to each step of cellular transformation and tumor progression
A.) Upon receiving an oncogenic insult (represented by a lightning bolt), e.g. Myc overexpression or PI3K hyperactivation, cells induce ribosome biogenesis and global protein synthesis that leads to increased cell size, coupled to cell division. Upon oncogenic stress, cells initiate a tumor suppressive response, associated with increased IRES mediated translation, leading to cell cycle arrest and senescence. In order to overcome the barrier of oncogene-induced senescence, cells acquire additional mutations (secondary hits, represented by lightning bolt). One mechanism for this is decreased translation from the CDK11/p58 IRES during mitosis, resulting from Myc hyperactivation, that leads to genome instability. B.) Once established, a primary tumor will undergo unrestrained growth, leading to a stress response associated with depletion of oxygen and essential nutrients from the core of the tumor. Lack of key nutrients, such as growth factors, often leads to increased apoptosis (skulls). Tumor cells upregulate cap-independent translation of anti-apoptotic factors (such as Bcl-2 and XIAP) as a mechanism to promote survival (blocked skulls). To bypass stress caused by low levels of oxygen in the tumor, cells induce cap-independent translation of neo-angiogenesis promoting factors such as VEGF.
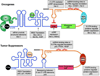
Figure 2. Oncogenes and Tumor Suppressors are translationally regulated through specific regulatory elements in their mRNAs
Depictions of cis-regulatory elements present in oncogenes (top) and tumor suppressor (bottom) mRNAs. Examples of regulated mRNAs are given (grey boxes). The cap binding protein eIF4E binds the 5’ cap of mRNAs. Increased eIF4E activity recruits eIF4G and the eIF4A helicase to unwind (green arrow) structured elements in the 5’UTR of poorly translated mRNAs, increasing the expression of many growth promoting and pro-survival genes, such as Mcl-1. Another group of structural elements that direct translational regulation are IRES elements. IRES elements promote cap-independent protein synthesis of both tumor suppressors (p53) and pro-survival factors (XIAP) during different stages of tumor development (see Figure 1). IRES Trans-Acting Factors (ITAFs) modulate translational regulation directed by specific IRES elements. Upstream Open Reading Frames (uORF) are present in select mRNAs and inhibit protein synthesis by preventing the ribosome from scanning to the start codon. Translation of growth promoting factors, such as Mdm2 and Her-2, is limited by uORFs. Additionally, a region in the 3’UTR of the Her-2 mRNA can inhibit uORF mediated repression of translation. 3’UTRs contain many distinct elements that interact with RNA binding proteins (RBPs) and miRNAs to promote or inhibit translation. One interesting example is the regulation of p53 translation mediated by base pairing interactions between elements in the 5’ and 3’ UTRs of the p53.
Similar articles
- Translational control in cancer etiology.
Ruggero D. Ruggero D. Cold Spring Harb Perspect Biol. 2013 Feb 1;5(2):a012336. doi: 10.1101/cshperspect.a012336. Cold Spring Harb Perspect Biol. 2013. PMID: 22767671 Free PMC article. Review. - The plasticity of mRNA translation during cancer progression and therapy resistance.
Fabbri L, Chakraborty A, Robert C, Vagner S. Fabbri L, et al. Nat Rev Cancer. 2021 Sep;21(9):558-577. doi: 10.1038/s41568-021-00380-y. Epub 2021 Aug 2. Nat Rev Cancer. 2021. PMID: 34341537 Review. - uORF-mediated translational control: recently elucidated mechanisms and implications in cancer.
Chen HH, Tarn WY. Chen HH, et al. RNA Biol. 2019 Oct;16(10):1327-1338. doi: 10.1080/15476286.2019.1632634. Epub 2019 Jun 24. RNA Biol. 2019. PMID: 31234713 Free PMC article. Review. - Translational control in cancer.
Silvera D, Formenti SC, Schneider RJ. Silvera D, et al. Nat Rev Cancer. 2010 Apr;10(4):254-66. doi: 10.1038/nrc2824. Nat Rev Cancer. 2010. PMID: 20332778 Review. - Translation initiation in cancer: a novel target for therapy.
Meric F, Hunt KK. Meric F, et al. Mol Cancer Ther. 2002 Sep;1(11):971-9. Mol Cancer Ther. 2002. PMID: 12481419 Review.
Cited by
- 4EBP-Dependent Signaling Supports West Nile Virus Growth and Protein Expression.
Shives KD, Massey AR, May NA, Morrison TE, Beckham JD. Shives KD, et al. Viruses. 2016 Oct 18;8(10):287. doi: 10.3390/v8100287. Viruses. 2016. PMID: 27763553 Free PMC article. - Ribosome biogenesis dysfunction leads to p53-mediated apoptosis and goblet cell differentiation of mouse intestinal stem/progenitor cells.
Stedman A, Beck-Cormier S, Le Bouteiller M, Raveux A, Vandormael-Pournin S, Coqueran S, Lejour V, Jarzebowski L, Toledo F, Robine S, Cohen-Tannoudji M. Stedman A, et al. Cell Death Differ. 2015 Nov;22(11):1865-76. doi: 10.1038/cdd.2015.57. Epub 2015 Jun 12. Cell Death Differ. 2015. PMID: 26068591 Free PMC article. - Automated high-throughput RNAi screening in human cells combined with reporter mRNA transfection to identify novel regulators of translation.
Casanova CM, Sehr P, Putzker K, Hentze MW, Neumann B, Duncan KE, Thoma C. Casanova CM, et al. PLoS One. 2012;7(9):e45943. doi: 10.1371/journal.pone.0045943. Epub 2012 Sep 27. PLoS One. 2012. PMID: 23029333 Free PMC article. - eIF4E-Dependent Translational Control: A Central Mechanism for Regulation of Pain Plasticity.
Uttam S, Wong C, Price TJ, Khoutorsky A. Uttam S, et al. Front Genet. 2018 Oct 24;9:470. doi: 10.3389/fgene.2018.00470. eCollection 2018. Front Genet. 2018. PMID: 30459806 Free PMC article. Review. - EIF3C-enhanced exosome secretion promotes angiogenesis and tumorigenesis of human hepatocellular carcinoma.
Lee HY, Chen CK, Ho CM, Lee SS, Chang CY, Chen KJ, Jou YS. Lee HY, et al. Oncotarget. 2018 Jan 11;9(17):13193-13205. doi: 10.18632/oncotarget.24149. eCollection 2018 Mar 2. Oncotarget. 2018. PMID: 29568350 Free PMC article.
References
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. - PubMed
- Ruggero D, Grisendi S, Piazza F, Rego E, Mari F, Rao PH, Cordon-Cardo C, Pandolfi PP. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. - PubMed
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. - PubMed
- Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010:1–12. This paper demonstrated a translational switch during OIS leading to increased IRES translation. This switch is impaired when ribosomes are not fully modified, leading to decreased p53 function in vivo.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources