PathSeq: software to identify or discover microbes by deep sequencing of human tissue - PubMed (original) (raw)
- PMID: 21552235
- PMCID: PMC3523678
- DOI: 10.1038/nbt.1868
PathSeq: software to identify or discover microbes by deep sequencing of human tissue
Aleksandar D Kostic et al. Nat Biotechnol. 2011 May.
No abstract available
Figures
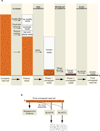
Figure 1. The PathSeq workflow
(a) Conceptual workflow of the subtractive phase of PathSeq. The size of the read set (orange bars) is proportional to the number of reads at the indicated step in a typical run of the method. The black dots in the bars represent pathogen-derived sequences which become progressively concentrated. The steps in this conceptual workflow have been reordered for concision (see Methods for actual ordering). (b) Conceptual workflow of the analytic phase of PathSeq. The asterisk indicates the unmapped readset that is carried over from the subtractive phase.
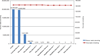
Figure 2. PathSeq performance on artificially generated sequence data
Reads were generated by sampling random 100-mer sequences from a human transcriptome database to produce 20 million reads, and from a set of twelve virus genomes each substitutionally mutated at twelve distinct rates, generating 144,000 reads (see Supplementary Fig. 1). The blue bars represent the number of human reads remaining after the indicated step in the PathSeq workflow, and the red squares connected by a line represent the remaining viral reads.
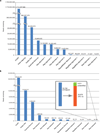
Figure 3. PathSeq performance on experimental sequence data
(a) We applied whole-genome sequencing data from a human ovarian tumor and (b) one lane of total-RNA transcriptome sequencing from HeLa cell lines to PathSeq. The inset in panel b shows that the 30,790 reads remaining after the subtractive phase of PathSeq are predominantly composed of HPV-18 sequences.
References
- Relman DA. Science. 1999;284:1308–1310. - PubMed
- Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Nat Genet. 2002;30:141–142. - PubMed
- Xu Y, et al. Genomics. 2003;81:329–335. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources