Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing - PubMed (original) (raw)
Comparative Study
Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing
Robert Willem van Nues et al. EMBO J. 2011.
Abstract
2'-O-methylation of eukaryotic ribosomal RNA (r)RNA, essential for ribosome function, is catalysed by box C/D small nucleolar (sno)RNPs. The RNA components of these complexes (snoRNAs) contain one or two guide sequences, which, through base-pairing, select the rRNA modification site. Adjacent to the guide sequences are protein-binding sites (the C/D or C'/D' motifs). Analysis of >2000 yeast box C/D snoRNAs identified additional conserved sequences in many snoRNAs that are complementary to regions adjacent to the rRNA methylation site. This 'extra base-pairing' was also found in many human box C/D snoRNAs and can stimulate methylation by up to five-fold. Sequence analysis, combined with RNA-protein crosslinking in Saccharomyces cerevisiae, identified highly divergent box C'/D' motifs that are bound by snoRNP proteins. In vivo rRNA methylation assays showed these to be active. Our data suggest roles for non-catalytic subunits (Nop56 and Nop58) in rRNA binding and support an asymmetric model for box C/D snoRNP organization. The study provides novel insights into the extent of the snoRNA-rRNA interactions required for efficient methylation and the structural organization of the snoRNPs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
Sequence alignments of box C/D snoRNAs. (A) Homologues for each of the S. cerevisiae box C/D snoRNAs were retrieved from the fungal genomic sequence databases and aligned. Two example alignments, using a limited subset of the sequences for snR74 and snR75, are shown. The sequence is shown 5′–3′ and the position of the box sequences are indicated, with the consensus sequence shown at the bottom. The rRNA target (3′–5′) is shown in white on a red background. The extra base-pairing target of snR75 is shown in white with a blue background. Identical sequences: white with a black background; conserved sequences: black with a grey background. Brackets indicate possible intra-molecular base-pairing. Scer: Saccharomyces cerevisiae; Cgla: Candida glabrata; Klac: Kluyveromyces lactis; Lelo: Lodderomyces elongisporus; Wano: Wickerhamomyces anomalus (Pichia anomala); Sjap: Schizosaccharomyces japonicas; Tree: Trichoderma reesei (Hypocrea jecorina); Tsti: Talaromyces stipitatus; Acla: Aspergillus clavatus; Nfis: Neosartorya fischeri; Cpos: Coccidioides posadasii; Pans: Podospora anserine. (B) The D′ and C′ sequences of the S. cerevisiae box C/D snoRNAs are shown. Insertions in the C′ boxes are indicated in red. The snoRNAs containing box C′/D′ motifs that do not appear to direct methylation are indicated in grey. (C) A schematic representation of the conservation of the sequences of the C, D, C′ and D′ boxes of the S. cerevisiae box C/D. The diagram was prepared using the WebLogo software (Crooks et al, 2004).
Figure 2
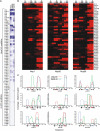
Asymmetric distribution of core snoRNP proteins on box C/D snoRNAs. (A) Box C/D snoRNAs are substantially enriched in core snoRNP protein CRAC Solexa data sets. Total hits for H/ACA and C/D snoRNAs were calculated in each data set, log transformed, clustered and displayed as a heatmap. Box C/D and H/ACA snoRNAs are indicated by brackets. (B) Heatmaps of average read densities along box C/D snoRNAs. The positions of C, D, C′ and D′ boxes (black), and the two guide regions (red), are indicated at the top. (C) Distribution of reads smaller than 20 nucleotides along individual snoRNAs is shown as plots. The Nop1 hits are shown in red, Nop56 hits are shown in green and the Nop58 hits are shown in blue. The number of hits for Nop58 for both snR57 and snR39 were below those recorded for the H/ACA snoRNA snR37, the baseline for these experiments, and were therefore represented using a dashed line. snoRNA genes and location of conserved sequences (blue), guide sequences (red) or C/D snoRNP boxes (black) are indicated below the x axis. Coverage (y axis) indicates a fraction and was calculated by dividing the number of times a nucleotide in a gene was found in a read by the total number of hits for the gene.
Figure 3
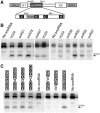
Methylation activity of C′/D′ motifs. (A) Schematic representation of the galactose-inducible snoRNA expression cassette. The positions of the GAL promoter (GALp), ADH terminator sequence (ADHt) and exons 1 and 2 of the actin gene (E1 and E2) are shown. The positions of the Nhe I and Mlu I restriction sites, used in the cloning of the various C′/D′ fragments, are indicated. The C′/D′ sequences cloned into this cassette are shown in Supplementary Figure S10. (B, C) snoRNAs containing wild-type and mutant C′ boxes (as indicated above each lane) were transformed into yeast cells. RNA was extracted from the cells and analysed by primer extension, using primer Map1316 (upper panel), to detect rRNA methylation, and by northern hybridization (Supplementary Figure S2) to detect the expression of the snoRNA. The position of the stop corresponding to methylation of the target nucleotide, S1316 in the 18S rRNA, is indicated on the right. The snoRNA containing the C′/D′ motif from hU24 was used in all experiments to enable the comparison of the relative methylation activity of the various C′/D′ motifs.
Figure 4
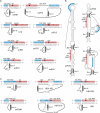
Extra conserved snoRNA sequences are complementary to rRNA target sites. (A) rRNA (upper) and snoRNA (lower) sequences, with both conventional guide-rRNA interactions (red) and novel extra base-pairing (blue) interactions for S. cerevisiae snoRNAs, are shown. Where sequences are shaded both red and blue, this indicates an overlap between the conventional and extra base-pairing. The D or D′ sequences are shown in white with a black background. (B) Schematic representations of S. cerevisiae snoRNA secondary structures with rRNA target sequence interactions. The regions base-paired to the guide and the extra base-pairing region are indicated using a red and blue background, respectively. Conserved boxes are indicated and the sequence shown in white on a black background. (C) Human snoRNA–rRNA interactions are schematically represented as in (A).
Figure 5
Extra base-pairing sequences are important for efficient methylation. (A) snR75 and snR76 interactions with the 25S rRNA are shown. The regions bound by the guide and extra base-pairing sequence are indicated using a red and blue background, respectively. Conserved boxes are indicated and shown in white on a black background. The sequence of the mutated extra base-pairing regions is shown in lower case. (B) A S. cerevisiae strain, in which the snR72–snR78 cluster was deleted, was transformed with plasmids expressing the snR72–snR78 cluster containing the wild-type (wt) or mutant (mut) snR75 and snR76 snoRNA-coding sequences or the vector alone (−). RNA was extracted from the cells and analysed by site-specific RNase H cleavage, to detect rRNA methylation, and by northern hybridization (Supplementary Figure S2), to detect the expression of the snoRNA. The cleaved RNAs were separated on a glyoxal/agarose gel, stained with ethidium bromide and visualized using a transilluminator. The positions of the full-length rRNAs and the 25S (arrows) and 18S (asterisk) cleavage products are indicated on the right. Reactions were performed in the presence (+) or absence (−) of RNase H as indicated. The oligonucleotides used for the analysis of the snR75 (upper panel) and snR76 (lower panel) modification sites are indicated on the left. (C) The region between the D′ and C′ boxes of the snR70 snoRNA was cloned into the artificial snoRNA (Figure 3A) to target the site S1315 in the 18S rRNA (snR70C′/D′). The extra guide region was then mutated (sequence shown in lower case) so that it was complementary to the region just upstream of the 18S rRNA target site. (D) Plasmids expressing the snoRNAs and a snoRNA containing the human U24 C′/D′ motif (targeting S1316) were transformed into yeast. RNA was extracted and analysed by primer extension using primer Map1316 and by northern blotting (Supplementary Figure S2). The positions of the stop corresponding to methylation of the target nucleotides, S1316 and S1315 in the 18S rRNA, are indicated on the right.
Figure 6
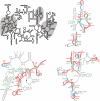
snoRNA base-pairing with the 25S rRNA. A line drawing of the secondary structure of the S. cerevisiae 25S and 5.8S rRNAs is shown at the top. The three regions containing modifications are shaded grey. The detailed secondary structures (obtained from
http://www.rna.ccbb.utexas.edu/
) of the three modified regions are also shown with the methylation (M) and pseudouridylation (Ψ) sites, and modifying snoRNAs, indicated in red and green, respectively. The methylation guide and extra base-pairing interaction sites in the rRNA are indicated by red and blue lines, respectively. Grey lines connect the lines for conventional guide and extra base-pairing interactions from one snoRNA.
Similar articles
- Unusual C΄/D΄ motifs enable box C/D snoRNPs to modify multiple sites in the same rRNA target region.
van Nues RW, Watkins NJ. van Nues RW, et al. Nucleic Acids Res. 2017 Feb 28;45(4):2016-2028. doi: 10.1093/nar/gkw842. Nucleic Acids Res. 2017. PMID: 28204564 Free PMC article. - A conserved Bcd1 interaction essential for box C/D snoRNP biogenesis.
Khoshnevis S, Dreggors RE, Hoffmann TFR, Ghalei H. Khoshnevis S, et al. J Biol Chem. 2019 Nov 29;294(48):18360-18371. doi: 10.1074/jbc.RA119.010222. Epub 2019 Sep 19. J Biol Chem. 2019. PMID: 31537647 Free PMC article. - Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly.
Rothé B, Manival X, Rolland N, Charron C, Senty-Ségault V, Branlant C, Charpentier B. Rothé B, et al. Nucleic Acids Res. 2017 Jul 7;45(12):7455-7473. doi: 10.1093/nar/gkx424. Nucleic Acids Res. 2017. PMID: 28505348 Free PMC article. - The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA.
Watkins NJ, Bohnsack MT. Watkins NJ, et al. Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):397-414. doi: 10.1002/wrna.117. Epub 2011 Nov 7. Wiley Interdiscip Rev RNA. 2012. PMID: 22065625 Review. - The multistructural forms of box C/D ribonucleoprotein particles.
Yu G, Zhao Y, Li H. Yu G, et al. RNA. 2018 Dec;24(12):1625-1633. doi: 10.1261/rna.068312.118. Epub 2018 Sep 25. RNA. 2018. PMID: 30254138 Free PMC article. Review.
Cited by
- Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits.
Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Böttcher B, Granneman S, Watkins NJ, Tollervey D. Lebaron S, et al. Nat Struct Mol Biol. 2012 Aug;19(8):744-53. doi: 10.1038/nsmb.2308. Epub 2012 Jul 1. Nat Struct Mol Biol. 2012. PMID: 22751017 Free PMC article. - NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation.
Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, Rodnina MV, Höbartner C, Bohnsack MT. Haag S, et al. EMBO J. 2016 Oct 4;35(19):2104-2119. doi: 10.15252/embj.201694885. Epub 2016 Aug 5. EMBO J. 2016. PMID: 27497299 Free PMC article. - Fractional 2'-O-methylation in the ribosomal RNA of Dictyostelium discoideum supports ribosome heterogeneity in Amoebozoa.
Diesend J, Birkedal U, Kjellin J, Zhang J, Jablonski KP, Söderbom F, Nielsen H, Hammann C. Diesend J, et al. Sci Rep. 2022 Feb 4;12(1):1952. doi: 10.1038/s41598-022-05447-w. Sci Rep. 2022. PMID: 35121764 Free PMC article. - Biology and applications of small nucleolar RNAs.
Bratkovič T, Rogelj B. Bratkovič T, et al. Cell Mol Life Sci. 2011 Dec;68(23):3843-51. doi: 10.1007/s00018-011-0762-y. Epub 2011 Jul 12. Cell Mol Life Sci. 2011. PMID: 21748470 Free PMC article. Review. - Csde1 binds transcripts involved in protein homeostasis and controls their expression in an erythroid cell line.
Moore KS, Yagci N, van Alphen F, Paolini NA, Horos R, Held NM, Houtkooper RH, van den Akker E, Meijer AB, 't Hoen PAC, von Lindern M. Moore KS, et al. Sci Rep. 2018 Feb 8;8(1):2628. doi: 10.1038/s41598-018-20518-7. Sci Rep. 2018. PMID: 29422612 Free PMC article.
References
- Decatur WA, Fournier MJ (2002) rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351 - PubMed
- Dennis PP, Omer A (2005) Small non-coding RNAs in Archaea. Curr Opin Microbiol 8: 685–694 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases