Non-coding RNAs as regulators of gene expression and epigenetics - PubMed (original) (raw)
Review
. 2011 Jun 1;90(3):430-40.
doi: 10.1093/cvr/cvr097. Epub 2011 May 9.
Affiliations
- PMID: 21558279
- PMCID: PMC3096308
- DOI: 10.1093/cvr/cvr097
Review
Non-coding RNAs as regulators of gene expression and epigenetics
Minna U Kaikkonen et al. Cardiovasc Res. 2011.
Abstract
Genome-wide studies have revealed that mammalian genomes are pervasively transcribed. This has led to the identification and isolation of novel classes of non-coding RNAs (ncRNAs) that influence gene expression by a variety of mechanisms. Here we review the characteristics and functions of regulatory ncRNAs in chromatin remodelling and at multiple levels of transcriptional and post-transcriptional regulation. We also describe the potential roles of ncRNAs in vascular biology and in mediating epigenetic modifications that might play roles in cardiovascular disease susceptibility. The emerging recognition of the diverse functions of ncRNAs in regulation of gene expression suggests that they may represent new targets for therapeutic intervention.
Figures
Figure 1
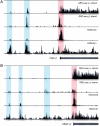
Potential promoter-associated RNAs and enhancer RNAs produced upstream of (A) fibroblast growth factor 2 (FGF-2) and (B) vascular endothelial growth factor C (VEGF-C) genes in human IMR90 cells. The promoter-associated RNAs (highlighted in red) colocalize with H3K4me3 histone mark, whereas the eRNAs (highlighted in blue) are revealed by their overlap with H3K4me1 mark. Regions upstream of transcription start site (TSS) for FGF-2 and VEGF-C are 50 and 100 kb, respectively. The _y_-axis indicates the number of sequencing tags.
Figure 2
Mechanisms for regulation of epigenetics and gene expression by non-coding RNAs. NcRNAs can function as modulators of epigenetics through (A through C) chromatin remodelling or regulate gene expression at (D through F) transcriptional or (G through I) post-transcriptional level. (A) A 5′ domain of HOTAIR binds polycomb repressive complex 2 (PRC2), whereas a 3′ domain of HOTAIR binds the LSD1/CoREST/REST complex. This allows HOTAIR to coordinate histone H3 lysine 27 methylation and lysine 4 demethylation at the HOXD locus in trans. (B) In cis recruitment of PRC2 by Xist antisense RNA and appearance of H3K27me3 along the inactive X chromosome are among the earliest events in X inactivation. Recruitment of PRC1-mediated H2AK119ub1 parallels the recruitment of PRC2. (C) Similarly, antisense non-coding RNA ANRIL represses the expression from INK4b/ARF/INK4a locus by recruiting and retaining PRC1 and PRC2 complexes in cis. (D) LncRNA transcribed from the minor promoter of dihydrofolate reductase (DHFR) froms a triplex together with the transcription factor TFIIB and the major promoter leading to the dissociation of the preinitiation complex. (E) Enhancer region (i and ii) of Dlx5/6 generates an lncRNA Evf-2 which forms a complex with homeodomain protein Dlx-2 to activate transcription. (F) Transcription of B2 and Alu RNAs is induced upon heat-shock. They inhibit mRNA synthesis by disrupting contacts between RNA polymerase II and promoter DNA. (G) Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are incorporated into RNA-induced silencing complexes (RISCs) that target specific mRNAs for cleavage, translational repression or destabilization depending on the extent of sequence complementarity. (H) Natural antisense transcript (NAT) prevents the binding of the spliceosome to the 5′UTR of the Zeb2 mRNA. This leads to retention in an intron containing internal ribosomal entry site (IRES), which is dispensable for the translation of Zeb2 protein. (I) The nuclear trafficking of nuclear factor of activated T cells (NFAT) is inhibited by the interaction of non-coding repressor of NFAT (NRON) with proteins of the importin-beta superfamily.
Similar articles
- RNA-mediated gene activation.
Jiao AL, Slack FJ. Jiao AL, et al. Epigenetics. 2014 Jan;9(1):27-36. doi: 10.4161/epi.26942. Epub 2013 Nov 1. Epigenetics. 2014. PMID: 24185374 Free PMC article. Review. - Emerging roles of non-coding RNAs in epigenetic regulation.
Chen J, Xue Y. Chen J, et al. Sci China Life Sci. 2016 Mar;59(3):227-35. doi: 10.1007/s11427-016-5010-0. Epub 2016 Jan 29. Sci China Life Sci. 2016. PMID: 26825947 Review. - Non-coding RNAs: regulators of disease.
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Taft RJ, et al. J Pathol. 2010 Jan;220(2):126-39. doi: 10.1002/path.2638. J Pathol. 2010. PMID: 19882673 Review. - Genome-wide expression of non-coding RNA and global chromatin modification.
Zhang R, Zhang L, Yu W. Zhang R, et al. Acta Biochim Biophys Sin (Shanghai). 2012 Jan;44(1):40-7. doi: 10.1093/abbs/gmr112. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22194012 Review. - Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs.
Malecová B, Morris KV. Malecová B, et al. Curr Opin Mol Ther. 2010 Apr;12(2):214-22. Curr Opin Mol Ther. 2010. PMID: 20373265 Free PMC article. Review.
Cited by
- Characteristic patterns of microRNA expression in human bladder cancer.
Zabolotneva AA, Zhavoronkov A, Garazha AV, Roumiantsev SA, Buzdin AA. Zabolotneva AA, et al. Front Genet. 2013 Jan 4;3:310. doi: 10.3389/fgene.2012.00310. eCollection 2012. Front Genet. 2013. PMID: 23316212 Free PMC article. - Micro-RNAs: Crossroads between the Exposure to Environmental Particulate Pollution and the Obstructive Pulmonary Disease.
Finicelli M, Squillaro T, Galderisi U, Peluso G. Finicelli M, et al. Int J Mol Sci. 2020 Sep 30;21(19):7221. doi: 10.3390/ijms21197221. Int J Mol Sci. 2020. PMID: 33007849 Free PMC article. Review. - Mitochondrial genomic variation drives differential nuclear gene expression in discrete regions of Drosophila gene and protein interaction networks.
Mossman JA, Biancani LM, Rand DM. Mossman JA, et al. BMC Genomics. 2019 Sep 2;20(1):691. doi: 10.1186/s12864-019-6061-y. BMC Genomics. 2019. PMID: 31477008 Free PMC article. - Emerging role of epigenetic regulations in periodontitis: a literature review.
Huang J, Zhou Y. Huang J, et al. Am J Transl Res. 2022 Apr 15;14(4):2162-2183. eCollection 2022. Am J Transl Res. 2022. PMID: 35559409 Free PMC article. Review. - Epigenetics in systemic lupus erythematosus.
Xiao G, Zuo X. Xiao G, et al. Biomed Rep. 2016 Feb;4(2):135-139. doi: 10.3892/br.2015.556. Epub 2015 Dec 11. Biomed Rep. 2016. PMID: 26893827 Free PMC article.
References
- The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. - PubMed
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. - PubMed