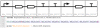Eugene--a domain specific language for specifying and constraining synthetic biological parts, devices, and systems - PubMed (original) (raw)
Eugene--a domain specific language for specifying and constraining synthetic biological parts, devices, and systems
Lesia Bilitchenko et al. PLoS One. 2011.
Abstract
Background: Synthetic biological systems are currently created by an ad-hoc, iterative process of specification, design, and assembly. These systems would greatly benefit from a more formalized and rigorous specification of the desired system components as well as constraints on their composition. Therefore, the creation of robust and efficient design flows and tools is imperative. We present a human readable language (Eugene) that allows for the specification of synthetic biological designs based on biological parts, as well as provides a very expressive constraint system to drive the automatic creation of composite Parts (Devices) from a collection of individual Parts.
Results: We illustrate Eugene's capabilities in three different areas: Device specification, design space exploration, and assembly and simulation integration. These results highlight Eugene's ability to create combinatorial design spaces and prune these spaces for simulation or physical assembly. Eugene creates functional designs quickly and cost-effectively.
Conclusions: Eugene is intended for forward engineering of DNA-based devices, and through its data types and execution semantics, reflects the desired abstraction hierarchy in synthetic biology. Eugene provides a powerful constraint system which can be used to drive the creation of new devices at runtime. It accomplishes all of this while being part of a larger tool chain which includes support for design, simulation, and physical device assembly.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
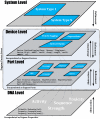
Figure 1. Encapsulation based synthetic biology design hierarchy.
Shown are the various layers of abstraction at which Eugene operates. DNA information forms the most basic unit on which everything else is built (e.g. the genetic code, as specified by bases G, A, T, and C). This is followed by Parts. Parts are non-reducible elements of genetic composition (e.g. promoters, ribosome binding sites, open reading frames, etc). Devices, which can contain one or more Parts, are the next level in the hierarchy. Finally, Devices are followed by a System view that contains collections of Devices. The traversal upward in the hierarchy represents an abstraction process while a downward traversal represents the refinement process. Eugene currently operates at the Part and Device levels via explicit Part and Device data types while encapsulating the DNA level as Eugene Properties.
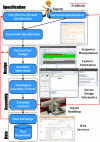
Figure 2. Eugene based synthetic biology design flow.
Shown here is the role that Specification, Design, Assembly, and Data can play in synthetic biology. In particular, we illustrate that Eugene is concerned with the activities at the specification level explicitly but at the same time it is designed in such a way that it develops designs that are amenable to other pieces of this design flow. Opportunities for the flow to provide feedback to earlier stages and perform iterative refinement are outlined in red.
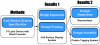
Figure 3. This paper is organized around three sections which reflect a Eugene design flow.
The Methods section provides an overview of how to use Eugene to create a Device using a T4 lysis Device from the MIT parts registry. The Results section illustrates design space exploration with a Cell Surface Display system from UC Berkeley's 2009 iGEM team. The Results section also details integration of Eugene with automated assembly in Clotho for a protein tagging system and simulation via SynBioSS for a repressilator design.
Figure 4. We illustrate both visually (SBOL visual; http://www.sbolstandard.org) and textually (Eugene code) an example Device (BBa_K112809) from the MIT registry of standardized parts.
Key to notice is the fact that the three included header files encapsulate much of the design effort leaving a single line to produce the composite Device.
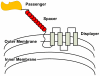
Figure 5. Illustration of the “cell surface display” Device case study.
Here are shown the three Part types (passengers, spaces, and displayers) which when combined into a Device made up the systems that we explored. As shown the displayer interacts with the outer membrane of the bacterial cell to display the passenger protein extracellularly. Table 2 in file Appendix S1 provides more information on this system.
Figure 6. Device exploration and pruning capabilities with Eugene.
This graph shows how the number of Devices created with Eugene can change with the addition of rule statements. The change in many cases can be quite dramatic with relatively few lines of code (new rules). For example with just two lines of code the initial design space explodes from two devices to 540. Then with 13 additional lines, it drops to 135 Devices. Finally, a design space of three Devices can be achieved by a total addition of 28 lines of code while still maintaining the original information to specify 540 Devices.
Figure 7. A heat map depicting the functionality of the cell surface display Devices, where the white constructs had the highest signal of functionality.
This data was used to determine which Devices could be considered functional and which were not. This analysis helped to drive the development of the Eugene code. The overlaid annotations reflect a reduced heat map. This shows how Devices can be removed with the targeted application of Eugene rules. The entire design space of 90 Devices is a reduction from the original heat map's 135 Devices. Each area is labeled with the rules that affect the creation of these Devices. Rules 1-4 deal with the removal of Devices while Rules 5–6 preserve the final three highly active Devices. The x-axis is displayer domain parts and the y-axis is protein/spacer combinations.
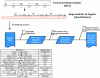
Figure 8. Illustration of an automated assembly flow beginning with a Eugene file for a protein tagging (PT) Device with nTag and cTag Parts.
This shows the eight Parts that make up the Device and the order in which the Parts must be assembled to have a functional Device. In the Eugene import process, the Devices of interest are captured with Eugene and processed by a Clotho App (e.g. Spectacles). Later the Device construction is planned for a specific assembly protocol with the creation of an assembly graph. In the final phase, the files for a liquid handling robot are created and fed to the platform doing the assembly.
Figure 9. High-level diagram of a repressilator as well as its Eugene implementation.
Here the relationship between LacI, TetR, and AraC and the promoters in the system is shown. This design was chosen since its behavior is well understood and can be easily decomposed into the individual Parts that make up the Device. The SynBioSS design flow with Eugene is also shown. Beginning with the Eugene XML produced by the Eugene interpreter, SimpleXML creates an array which holds the data from Eugene. After a reorganization process the data can now be transformed by SynBioSS into a reaction network in SBML or NetCDF which can then be simulated. Sample of the reaction network generated by SynBioSS Designer is also provided. These reactions describe the unregulated expression of TetR, as well as its dimerization and degradation. All rate laws are elementary and all kinetic data is in SI units unless otherwise noted. Asterisks indicate gamma-distributed reactions.
Similar articles
- The Eugene language for synthetic biology.
Bilitchenko L, Liu A, Densmore D. Bilitchenko L, et al. Methods Enzymol. 2011;498:153-72. doi: 10.1016/B978-0-12-385120-8.00007-3. Methods Enzymol. 2011. PMID: 21601677 - Engineering Genomes with Genotype Specification Language.
Wilson EH, Macklin C, Platt D. Wilson EH, et al. Methods Mol Biol. 2018;1772:373-398. doi: 10.1007/978-1-4939-7795-6_21. Methods Mol Biol. 2018. PMID: 29754240 - Specifying Combinatorial Designs with the Synthetic Biology Open Language (SBOL).
Roehner N, Bartley B, Beal J, McLaughlin J, Pocock M, Zhang M, Zundel Z, Myers CJ. Roehner N, et al. ACS Synth Biol. 2019 Jul 19;8(7):1519-1523. doi: 10.1021/acssynbio.9b00092. Epub 2019 Jul 1. ACS Synth Biol. 2019. PMID: 31260271 - In silico implementation of synthetic gene networks.
Marchisio MA. Marchisio MA. Methods Mol Biol. 2012;813:3-21. doi: 10.1007/978-1-61779-412-4_1. Methods Mol Biol. 2012. PMID: 22083733 Review. - A standard-enabled workflow for synthetic biology.
Myers CJ, Beal J, Gorochowski TE, Kuwahara H, Madsen C, McLaughlin JA, Mısırlı G, Nguyen T, Oberortner E, Samineni M, Wipat A, Zhang M, Zundel Z. Myers CJ, et al. Biochem Soc Trans. 2017 Jun 15;45(3):793-803. doi: 10.1042/BST20160347. Biochem Soc Trans. 2017. PMID: 28620041 Review.
Cited by
- Genetic design automation: engineering fantasy or scientific renewal?
Lux MW, Bramlett BW, Ball DA, Peccoud J. Lux MW, et al. Trends Biotechnol. 2012 Feb;30(2):120-6. doi: 10.1016/j.tibtech.2011.09.001. Epub 2011 Oct 14. Trends Biotechnol. 2012. PMID: 22001068 Free PMC article. Review. - Interactive assembly algorithms for molecular cloning.
Appleton E, Tao J, Haddock T, Densmore D. Appleton E, et al. Nat Methods. 2014 Jun;11(6):657-62. doi: 10.1038/nmeth.2939. Epub 2014 Apr 28. Nat Methods. 2014. PMID: 24776633 - Modeling and simulation of biological systems using SPICE language.
Madec M, Lallement C, Haiech J. Madec M, et al. PLoS One. 2017 Aug 7;12(8):e0182385. doi: 10.1371/journal.pone.0182385. eCollection 2017. PLoS One. 2017. PMID: 28787027 Free PMC article. - The PLOS ONE synthetic biology collection: six years and counting.
Peccoud J, Isalan M. Peccoud J, et al. PLoS One. 2012;7(8):e43231. doi: 10.1371/journal.pone.0043231. Epub 2012 Aug 15. PLoS One. 2012. PMID: 22916228 Free PMC article. Review. - Towards a behavioral-matching based compilation of synthetic biology functions.
Basso-Blandin A, Delaplace F. Basso-Blandin A, et al. Acta Biotheor. 2015 Sep;63(3):325-39. doi: 10.1007/s10441-015-9265-9. Epub 2015 Jul 5. Acta Biotheor. 2015. PMID: 26141968 Free PMC article.
References
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. - PubMed
- Lucks JB, Qi L, Whitaker WR, Arkin AP. Toward scalable parts families for predictable design of biological circuits. Curr Opin Microbiol. 2008;11:567–573. - PubMed
- Arkin A. Setting the standard in synthetic biology. Nat Biotechnol. 2008;26:771–774. - PubMed
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. - PubMed
- Stevenson D. A Proposed Standard for Binary Floating-Point Arithmetic. Computer. 1981;14:51–62.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources