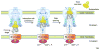Secretins: dynamic channels for protein transport across membranes - PubMed (original) (raw)
Review
Secretins: dynamic channels for protein transport across membranes
Konstantin V Korotkov et al. Trends Biochem Sci. 2011 Aug.
Abstract
Secretins form megadalton bacterial-membrane channels in at least four sophisticated multiprotein systems that are crucial for translocation of proteins and assembled fibers across the outer membrane of many species of bacteria. Secretin subunits contain multiple domains, which interact with numerous other proteins, including pilotins, secretion-system partner proteins, and exoproteins. Our understanding of the structure of secretins is rapidly progressing, and it is now recognized that features common to all secretins include a cylindrical arrangement of 12-15 subunits, a large periplasmic vestibule with a wide opening at one end and a periplasmic gate at the other. Secretins might also play a key role in the biogenesis of their cognate secretion systems.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Figure 1. Secretins in Gram-negative bacteria
Schematic view of the type II and type III secretion systems, type IV pili system and bacteriophage assembly system. The secretin is the major outer membrane component of all these systems. The insertion of secretins into the outer membrane is often assisted by specific lipoproteins called pilotins. The T2SS secretes exoproteins from the periplasm to the extracellular space in the folded form. The T2SS pseudopilus is formed by multiple pseudopilin subunits; the pseudopilus is thought to act as a piston and/or plug during the secretion process (Box 1). The T4PS is related to the T2SS in several architectural and functional aspects, but a key difference is that the pilus extends outside the bacterial surface. The T3SSs transport effectors directly to the eukaryotic cytoplasm or membrane via a hollow needle. The inner membrane complexes of the T2SS, T4PS and the T3SS are composed of multiple proteins that include at least one ATPase involved in providing energy for secretion or pilus extension/retraction processes. The filamentous phage assembly system is composed of a secretin and two inner membrane proteins. E, extracellular space; OM, outer membrane; P, periplasm; IM, inner membrane; C, cytoplasm.
Figure 2. Secretin domains – modular organization and structures
(a) Domain composition of secretins. Secretins are synthesized as precursors with N-terminal signal sequences recognized and cleaved off by a signal peptidase (grey) or a prolipoprotein signal peptidase (yellow). In the latter case, secretins are lipoproteins with acylated N-terminal Cys residues. The C-terminal secretin core homology domain (light blue; Pfam family PF00263 [75]) contains putative amphipathic transmembrane β-strands. The N-terminal N0 domain (purple; Pfam PF07660) is followed by one or several homologous repeat domains (light green; Pfam PF03958) termed N1–N3 depending on the number of repeats; some N3 domains have long loop insertions. The T4bPS secretins require small periplasmic proteins for stability and multimerization [65, 66] that also have putative N0 domains. Some secretins require specific lipoproteins, known as pilotins, for correct outer membrane targeting and contain C-terminal pilotin-interaction domains (dark blue), called S domains in the T2SS secretins. The pilotin-interaction domains are not related in sequence, which reflects the diversity of cognate pilotins. Some secretins contain domains of unknown topology (white). (b) Crystal structures of secretin domains. The N0-N1-N2 domains structure of ETEC GspD [15] (PDB 3EZJ) and the N0-N1 domains structure of EPEC EscC [16] (PDB 3GR5) are superimposed and shown in the same orientation relative to the N1 domains. The core of the N0 domain (purple) has a βαββαββ fold that is structurally related to the signaling domain of the TonB-dependent outer membrane receptors [18], a lipoprotein DotD from the Legionella pneumophila type IVb secretion system [20], a domain of protein VgrG from the E. coli type VI secretion system [19], and a domain of protein gp27 from T4-related bacteriophages [17]. As expected from sequence homology, the repeat N1 and N2 domains (light green) have similar βαββα folds; the first helix in the N1 domain is a tandem of 310 and α helices. The fold of N1 domain is different from N0, but structurally related to the eukaryotic type I KH (hnRNP K homology) domain [76], which is also found in several ring-forming inner membrane T3SS proteins: EPEC EscJ [21], S. typhimurium PrgH [16] and InvA [22, 23]. Although the structures of individual N0 and N1 domains of GspD and EscC superimpose well, the relative orientation of these domains and the N0–N1 contact interface is different in the T2SS and T3SS secretins. In the ETEC GspD structure, the N2 domain connects to the N0–N1 lobe via a potentially flexible linker. The relative orientation of the N2 domain with respect to the N0-N1 lobe is stabilized by crystal contacts and interactions with a nanobody (not shown) and is presumed to differ from the orientation in the secretin multimer [15].
Figure 3. Electron microscopy structures of secretins
(a) Cryo-EM reconstruction of V. cholerae T2SS secretin GspDEpsD [25] (EMDB 1763). In side view, three domains are identified from top to bottom: the extracellular cap, the outer membrane domain and the periplasmic domain. In a cross-section, the channel reveals an extracellular gate, an extracellular chamber, a periplasmic gate and a periplasmic vestibule with a constriction (yellow). (b) The secretin architecture is conserved in different secretion systems. (i): fitting of 12-fold symmetrical ring models of the N-terminal periplasmic domains (N0 domain in purple, N1–N3 domains in light green) into the GspDEpsD density map [25]. The N0 and N1 domains ring is anchored at the bottom of the map. The N2 domain ring fits into the central periplasmic domain density and the N3 domain ring into the periplasmic constriction. (ii): EM reconstruction of S. typhimurium T3SS base complex in the closed state [30] (EMDB 1224). OR – outer ring, IR – inner ring. (iii): EM reconstruction of the S. flexneri T3SS needle complex in the open state [31] (EMDB 1617). OMR – outer membrane ring, IMR – inner membrane ring. The secretin (blue) occupies the top part of the T3SS reconstructions above the inner membrane complex – (ii) and (iii). V. cholerae GspDEpsD secretin is in a closed state; compare (i) and (ii). The modeled locations of N0 (purple solid ovals) and N1 (light green solid ovals) domain rings in the T3SS secretins are shown [33, 41], as well as putative locations of the N3 (light green open ovals) domain ring in the constriction site (upper neck or OMR3) based on the corresponding N3 domain ring fit in the T2SS secretin. (c) Cryo-EM reconstruction of S. typhimurium T3SS secretin InvG as part of the needle complex [32]. (i): fitting of the periplasmic domains of InvG (N0 domain in purple, N1 domain in light green) into the density map (EMDB 1875). Inner membrane part of the needle complex is omitted for clarity. (ii)–(iii): tilted and top views of the C15 map (EMDB 1871) corresponding to the neck region of InvG secretin with fitting of the periplasmic domains (PDB 2Y9K). (d) Experimental visualization of the secretin in the T3SS complex. Class averages of the S. typhimurium T3SS needle complexes purified from wild type (i) and ΔinvG secretin mutant (ii) strains. (iii) The density difference between averaged images of wild type and ΔinvG complexes [(i) − (ii) = (iii)] indicates the position of the secretin in the complex. Reproduced from [33] under the terms of the Creative Commons Attribution License. Copyright: © 2010 Schraidt et al. (e) Electron microscopy analysis of the T. thermophilus T4PS secretin PilQ. A class average of purified PilQ shows a 150 Å wide and 340 Å long particle with features similar to the T2SS and T3SS secretins (compare with the side view (a) and (b, ii)) [29]. The upper (outer membrane) part of the PilQ particle reveals a bisecting density corresponding to the periplasmic gate. The conical lower (periplasmic) part of PilQ consists of six concentric rings that likely correspond to the N-terminal secretin domains; sequence and fold-recognition analysis of PilQ by the Pcons server [63] indicates the presence of an N0 domain and five putative N1-like domains. Reproduced with permission from [29]. Copyright: © 2011 the American Society for Biochemistry and Molecular Biology.
Figure 4. Structures of pilotins
These structures are remarkably dissimilar in spite of a common function. (a) The structure of the S. flexneri T3SS pilotin MxiM (green) in complex with the C-terminal fragment of the secretin MxiD (dark blue) [51] (PDB 2JW1). The ‘cracked β barrel’ of MxiM forms a cleft at the top for binding of a short α helix of MxiD. (b) The structure of the N. meningitidis T4PS pilotin PilW (orange) [59] (PDB 2VQ2) is composed of six tetratricopeptide repeat (TPR) motifs arranged as a super-helix. Structures of a homologous P. aeruginosa pilotin PilF (26% sequence identity with N. meningitidis PilW) show a similar arrangement of TPR motifs [56, 58].
Figure I
Mechanism of the T2SS. Modified and reproduced with permission from [25]. Copyright: © 2010 Reichow et al.
Similar articles
- Assembly and targeting of secretins in the bacterial outer membrane.
Natarajan J, Singh N, Rapaport D. Natarajan J, et al. Int J Med Microbiol. 2019 Nov;309(7):151322. doi: 10.1016/j.ijmm.2019.06.002. Epub 2019 Jun 19. Int J Med Microbiol. 2019. PMID: 31262642 Review. - Structural lessons on bacterial secretins.
Barbat B, Douzi B, Voulhoux R. Barbat B, et al. Biochimie. 2023 Feb;205:110-116. doi: 10.1016/j.biochi.2022.08.019. Epub 2022 Sep 9. Biochimie. 2023. PMID: 36096236 Review. - Bacterial secretins: Mechanisms of assembly and membrane targeting.
Silva YRO, Contreras-Martel C, Macheboeuf P, Dessen A. Silva YRO, et al. Protein Sci. 2020 Apr;29(4):893-904. doi: 10.1002/pro.3835. Epub 2020 Feb 19. Protein Sci. 2020. PMID: 32020694 Free PMC article. Review. - Scaffolding Protein GspB/OutB Facilitates Assembly of the Dickeya dadantii Type 2 Secretion System by Anchoring the Outer Membrane Secretin Pore to the Inner Membrane and to the Peptidoglycan Cell Wall.
Zhang S, Gu S, Rycroft P, Ruaudel F, Delolme F, Robert X, Ballut L, Pickersgill RW, Shevchik VE. Zhang S, et al. mBio. 2022 Jun 28;13(3):e0025322. doi: 10.1128/mbio.00253-22. Epub 2022 May 12. mBio. 2022. PMID: 35546537 Free PMC article. - Decoding the roles of pilotins and accessory proteins in secretin escort services.
Koo J, Burrows LL, Howell PL. Koo J, et al. FEMS Microbiol Lett. 2012 Mar;328(1):1-12. doi: 10.1111/j.1574-6968.2011.02464.x. Epub 2011 Dec 8. FEMS Microbiol Lett. 2012. PMID: 22098485 Review.
Cited by
- Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity.
Lu C, Turley S, Marionni ST, Park YJ, Lee KK, Patrick M, Shah R, Sandkvist M, Bush MF, Hol WG. Lu C, et al. Structure. 2013 Sep 3;21(9):1707-17. doi: 10.1016/j.str.2013.06.027. Epub 2013 Aug 15. Structure. 2013. PMID: 23954505 Free PMC article. - Transport proteins promoting Escherichia coli pathogenesis.
Tang F, Saier MH Jr. Tang F, et al. Microb Pathog. 2014 Jun-Jul;71-72:41-55. doi: 10.1016/j.micpath.2014.03.008. Epub 2014 Apr 18. Microb Pathog. 2014. PMID: 24747185 Free PMC article. - Structural insights into the secretin translocation channel in the type II secretion system.
Yan Z, Yin M, Xu D, Zhu Y, Li X. Yan Z, et al. Nat Struct Mol Biol. 2017 Feb;24(2):177-183. doi: 10.1038/nsmb.3350. Epub 2017 Jan 9. Nat Struct Mol Biol. 2017. PMID: 28067918 - A dodecameric ring-like structure of the N0 domain of the type II secretin from enterotoxigenic Escherichia coli.
Korotkov KV, Delarosa JR, Hol WGJ. Korotkov KV, et al. J Struct Biol. 2013 Sep;183(3):354-362. doi: 10.1016/j.jsb.2013.06.013. Epub 2013 Jun 29. J Struct Biol. 2013. PMID: 23820381 Free PMC article. - Novel Role for PilNO in Type IV Pilus Retraction Revealed by Alignment Subcomplex Mutations.
Leighton TL, Dayalani N, Sampaleanu LM, Howell PL, Burrows LL. Leighton TL, et al. J Bacteriol. 2015 Jul;197(13):2229-2238. doi: 10.1128/JB.00220-15. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917913 Free PMC article.
References
- Wooldridge K. Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. Caister Academic Press; 2009.
- Johnson TL, et al. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. - PubMed
- Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68:827–837. - PubMed
- Kirn TJ, et al. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol. 2003;49:81–92. - PubMed
- Hager AJ, et al. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol. 2006;62:227–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI034501-14/AI/NIAID NIH HHS/United States
- R01 AI034501-17/AI/NIAID NIH HHS/United States
- R01 AI034501-12/AI/NIAID NIH HHS/United States
- R01 AI034501/AI/NIAID NIH HHS/United States
- R01 AI034501-13/AI/NIAID NIH HHS/United States
- R01 AI034501-16A2/AI/NIAID NIH HHS/United States
- R01 AI034501-18/AI/NIAID NIH HHS/United States
- R56 AI034501/AI/NIAID NIH HHS/United States
- R01 AI034501-11/AI/NIAID NIH HHS/United States
- R01AI34501/AI/NIAID NIH HHS/United States
- R01 AI034501-15/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases