Paused RNA polymerase II as a developmental checkpoint - PubMed (original) (raw)
Review
Paused RNA polymerase II as a developmental checkpoint
Michael Levine. Cell. 2011.
Abstract
The textbook view of gene activation is that the rate-limiting step is the interaction of RNA polymerase II (Pol II) with the gene's promoter. However, studies in a variety of systems, including human embryonic stem cells and the early Drosophila embryo, have begun to challenge this view. There is increasing evidence that differential gene expression often depends on the regulation of transcription elongation via the release of Pol II from the proximal promoter. I review the implications of this mechanism of gene activation with respect to the orderly unfolding of complex gene networks governing animal development.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
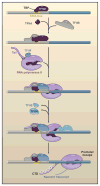
Figure 1. The First Steps in Transcription Activation
A transcription factor, such as TBP/TFIID, binds to specific promoter elements, including TATA. This leads to the recruitment of additional general transcription factors, including TFIIA, TFIIB, TFIIF, and ultimately, RNA polymerase II (Pol II). The initial binding is unstable, given that the promoter complex is in a closed conformation. Recruitment of TFIIH leads to the formation of an open complex and the onset of transcription. Stable transcription depends on the phosphorylation of the Pol II C-terminal region (CTD), which fosters promoter escape. Adapted from Gilmour (2009).
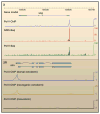
Figure 2. RNA Polymerase II Binding at Developmental Patterning Genes
RNA polymerase II (Pol II) binding at the proximal promoter regions of developmental patterning genes, including sog (A), which encodes a bone morphogenetic protein inhibitor, and sim (B), which specifies the ventral midline of the central nervous system. (A) Pol II chromatin immunoprecipitation (ChIP)-chip, genome-wide run on assays (Gro-Seq) and ChIP-Seq assays using extracts from 2–4 hr mutant embryos that lack a dorsal nuclear gradient and contain only dorsal ectoderm (Zeitlinger et al., 2007). Although sog is silent in these embryos, Pol II is clearly bound to the promoter region of the gene. (B) An ~10 kb region of the Drosophila genome, which contains the linked sim and timeout genes. ChIP-chip assays identified Pol II binding in the promoter region of sim, but not timeout, in early, 2–4 hr embryos. Pol II binding is seen at the sim promoter in pipe and rm9/rm10 mutants, which produce only dorsal ectoderm and neurogenic ectoderm, respectively. No binding is observed in Toll10b mutants, which produce only mesoderm (Zeitlinger et al., 2007).
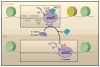
Figure 3. Paused Pol II and Its Release from the Proximal Promoter
The top panel shows a promoter DNA template with paused RNA polymerse II (Pol II). The promoter region contains sequence elements that foster binding and activation of Pol II, including GAGA, TATA, initiator, and downstream promoter element/pause button (DPE/PB) motifs. Pol II is typically paused just downstream of the DPE region. It has undergone promoter escape and contains phosphorylation of serine 5 (Ser5) in the C-terminal domain (CTD). DSIF (5,6-dichloro-1-β-D-ribofuranoxylbenzimidazole sensivitiy-inducing factor) and NELF (negative elongation factor) help to arrest Pol II by binding to the nascent transcript (typically 30–50 nucleotides in length). Recruitment of P-TEFb (positive transcription elongation factor b) causes the release of NELF and the phosphorylation of Ser2 in the CTD, resulting in Pol II procession into the main body of the gene.
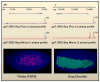
Figure 4. Activation of Paused and Nonpaused Genes
(Left) The Drosophila thisbe gene lacks binding of RNA polymerase II (Pol II), based on both chromatin immunoprecipitation assays (not shown) and Gro-Seq assays (shown). The gene exhibits a stochastic pattern of activation in the early embryo, with nascent transcripts detected in only about half of all nuclei that will ultimately express the gene. (Right) In contrast, the sog gene contains paused Pol II and exhibits a synchronous pattern of activation.
Similar articles
- Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo.
Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, Levine MS. Lagha M, et al. Cell. 2013 May 23;153(5):976-87. doi: 10.1016/j.cell.2013.04.045. Cell. 2013. PMID: 23706736 Free PMC article. - Promoter elements associated with RNA Pol II stalling in the Drosophila embryo.
Hendrix DA, Hong JW, Zeitlinger J, Rokhsar DS, Levine MS. Hendrix DA, et al. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7762-7. doi: 10.1073/pnas.0802406105. Epub 2008 May 27. Proc Natl Acad Sci U S A. 2008. PMID: 18505835 Free PMC article. - Promoter proximal pausing on genes in metazoans.
Gilmour DS. Gilmour DS. Chromosoma. 2009 Feb;118(1):1-10. doi: 10.1007/s00412-008-0182-4. Epub 2008 Oct 2. Chromosoma. 2009. PMID: 18830703 Review. - RNA polymerase II pausing during development.
Gaertner B, Zeitlinger J. Gaertner B, et al. Development. 2014 Mar;141(6):1179-83. doi: 10.1242/dev.088492. Development. 2014. PMID: 24595285 Free PMC article. Review. - PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II.
Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, Shiekhattar R, Shilatifard A. Chen FX, et al. Cell. 2015 Aug 27;162(5):1003-15. doi: 10.1016/j.cell.2015.07.042. Epub 2015 Aug 13. Cell. 2015. PMID: 26279188 Free PMC article.
Cited by
- A lineage-specific nascent RNA assay unveils principles of gene regulation in tissue biology.
Chovatiya G, Wang AB, Versluis P, Bai CK, Huang SY, DeBerardine M, Ray J, Ozer A, Lis JT, Tumbar T. Chovatiya G, et al. bioRxiv [Preprint]. 2024 Oct 18:2024.10.15.618417. doi: 10.1101/2024.10.15.618417. bioRxiv. 2024. PMID: 39464031 Free PMC article. Preprint. - PARP-1 is a transcriptional rheostat of metabolic and bivalent genes during development.
Bamgbose G, Tulin A. Bamgbose G, et al. Life Sci Alliance. 2023 Nov 27;7(2):e202302369. doi: 10.26508/lsa.202302369. Print 2024 Feb. Life Sci Alliance. 2023. PMID: 38012002 Free PMC article. - Enforcing the pause: transcription factor Sp3 limits productive elongation by RNA polymerase II.
Valin A, Gill G. Valin A, et al. Cell Cycle. 2013 Jun 15;12(12):1828-34. doi: 10.4161/cc.24992. Epub 2013 May 15. Cell Cycle. 2013. PMID: 23676218 Free PMC article. - Defective Hfp-dependent transcriptional repression of dMYC is fundamental to tissue overgrowth in Drosophila XPB models.
Lee JE, Mitchell NC, Zaytseva O, Chahal A, Mendis P, Cartier-Michaud A, Parsons LM, Poortinga G, Levens DL, Hannan RD, Quinn LM. Lee JE, et al. Nat Commun. 2015 Jun 15;6:7404. doi: 10.1038/ncomms8404. Nat Commun. 2015. PMID: 26074141 Free PMC article. - Genome-wide CRISPR knockout screen identifies ZNF304 as a silencer of HIV transcription that promotes viral latency.
Krasnopolsky S, Kuzmina A, Taube R. Krasnopolsky S, et al. PLoS Pathog. 2020 Sep 21;16(9):e1008834. doi: 10.1371/journal.ppat.1008834. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32956422 Free PMC article.
References
- Arndt KM, Kane CM. Running with RNA polymerase: eukaryotic transcript elongation. Trends Genet. 2003;19:543–550. - PubMed
- Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. - PubMed
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases