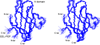Advanced glycation end product recognition by the receptor for AGEs - PubMed (original) (raw)
Advanced glycation end product recognition by the receptor for AGEs
Jing Xue et al. Structure. 2011.
Abstract
Nonenzymatic protein glycation results in the formation of advanced glycation end products (AGEs) that are implicated in the pathology of diabetes, chronic inflammation, Alzheimer's disease, and cancer. AGEs mediate their effects primarily through a receptor-dependent pathway in which AGEs bind to a specific cell surface associated receptor, the Receptor for AGEs (RAGE). N(ɛ)-carboxy-methyl-lysine (CML) and N(ɛ)-carboxy-ethyl-lysine (CEL), constitute two of the major AGE structures found in tissue and blood plasma, and are physiological ligands of RAGE. The solution structure of a CEL-containing peptide-RAGE V domain complex reveals that the carboxyethyl moiety fits inside a positively charged cavity of the V domain. Peptide backbone atoms make specific contacts with the V domain. The geometry of the bound CEL peptide is compatible with many CML (CEL)-modified sites found in plasma proteins. The structure explains how such patterned ligands as CML (CEL)-proteins bind to RAGE and contribute to RAGE signaling.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Figure 1
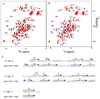
CEL (CML) containing peptides can be produced by both glycation and chemical synthesis. (A) Fructosyllysine, CML and CEL are the products of early and advanced glycoxidation of sugars. (B) Synthetic CEL-peptide, DEF(CEL)ADE, contains a correct chemical structure of _N_ε-carboxy-ethyl-lysine. To characterize CEL-peptide, we collected two 2D homonuclear NMR experiments, 1H,1H TOCSY and 1H, 1H ROESY. The 1H, 1H TOCSY strip shows through bond correlation between the amide proton (8ppm) and side chain protons Hα (4.1 ppm), Hβ (1.7 ppm), Hγ (1.2 ppm), Hδ (1.5 ppm), and Hε (2.85 ppm) of CEL. The 1H, 1H ROESY strip shows through space correlations between Hε and carboxyethyl protons CH(CH3)-COOH (3.8 ppm) and CH(_CH_3)-COOH (1.7 ppm) of CEL, confirming the presence of a proper chemical structure of _N_ε-carboxy-ethyl-lysine. See also Figure S1.
Figure 2
RAGE V domain does not discriminate between CML and CEL containing peptides. (A) Overlay of 15N-HSQC of free (black) and CML-PEP bound (red) V domain. (B) Overlay of 15N-HSQC of free (black) and CEL-PEP bound (red) V domain. (C) Sequence alignment of V domain and a V-type domain from a heavy chain antibody IgG1 P20.1. Amino acid residues involved in CML (CEL) binding are in red. CDR1 and CDR2, the hypervariable regions of IgG1 P20.1, are in blue. Secondary structure elements are shown above the sequences. See also Figure S3.
Figure 3
Stereoview of the overlay of 25 lowest energy CEL-PEP V domain backbone traces (PDB code 2L7U). N- and C-termini of the V domain and CEL-PEP are indicated. Figure is prepared by using Molmol (Koradi et al, 1996). See also Table S1.
Figure 4
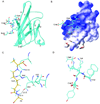
Solution structure of the CEL-PEP-V domain complex. (A) Structure of CEL-PEP bound to V domain. V domain is shown in ribbon representation. Elements of secondary structure are labeled following the immunoglobulin convention(Bork et al., 1994). (B) Electrostatic potential is mapped onto the molecular surface of the V domain. Positively and negatively charged surfaces are indicated in blue and red, respectively. (C) V domain amino acid residues located within 5 Å from the CEL moiety of CEL-PEP. Putative hydrogen bond between the backbone amide proton of Ala5 and the side chain carbonyl group of Asn112 is indicated. Carbon atoms of CEL-PEP and V domain are in yellow and cyan, respectively. (D) Structural alignment of CEL-PEP with three short segments from human serum albumin (HSA) containing lysines(Wa et al., 2007), 11-FKD, 56-AKT, and 261-AKY. The lysines in these sequences were shown to be glycated under elevated concentrations of D-glucose. See also Figure S4 and Table S2.
Figure 5
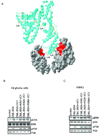
Mutants of the VC1 domains of RAGE fail to suppress CML-BSA induced RAGE signaling. (A) Cartoon model of how RAGE dimerization promotes V domain binding to multiple CML moieties on CML-BSA (ribbon). The molecular surface of the V domain involved in CML(CEL) binding is in red. Only two V domains are shown. Lysines of BSA, which may undergo glycation are in yellow. The cartoon model was prepared by using SWISS-PDB Viewer (Guex et al, 1997). (B, C) Single K52A and R98A, and double K52A, R98A (KRA) mutants of the VC1 domains do not interfere with CML-BSA induced RAGE signaling in both C6 rat glioma (b) and mouse VSMC cells (c). See also Figure S5.
Comment in
- How to create a specific recognition for an unspecific interaction.
Dötsch V. Dötsch V. Structure. 2011 May 11;19(5):601-2. doi: 10.1016/j.str.2011.04.003. Structure. 2011. PMID: 21565694
Similar articles
- Advanced glycation end product ligands for the receptor for advanced glycation end products: biochemical characterization and formation kinetics.
Valencia JV, Weldon SC, Quinn D, Kiers GH, DeGroot J, TeKoppele JM, Hughes TE. Valencia JV, et al. Anal Biochem. 2004 Jan 1;324(1):68-78. doi: 10.1016/j.ab.2003.09.013. Anal Biochem. 2004. PMID: 14654047 - The receptor for advanced glycation end products (RAGE) specifically recognizes methylglyoxal-derived AGEs.
Xue J, Ray R, Singer D, Böhme D, Burz DS, Rai V, Hoffmann R, Shekhtman A. Xue J, et al. Biochemistry. 2014 May 27;53(20):3327-35. doi: 10.1021/bi500046t. Epub 2014 May 13. Biochemistry. 2014. PMID: 24824951 Free PMC article. - A capture method based on the VC1 domain reveals new binding properties of the human receptor for advanced glycation end products (RAGE).
Degani G, Altomare AA, Colzani M, Martino C, Mazzolari A, Fritz G, Vistoli G, Popolo L, Aldini G. Degani G, et al. Redox Biol. 2017 Apr;11:275-285. doi: 10.1016/j.redox.2016.12.017. Epub 2016 Dec 18. Redox Biol. 2017. PMID: 28013188 Free PMC article. - The biology of the receptor for advanced glycation end products and its ligands.
Schmidt AM, Yan SD, Yan SF, Stern DM. Schmidt AM, et al. Biochim Biophys Acta. 2000 Dec 20;1498(2-3):99-111. doi: 10.1016/s0167-4889(00)00087-2. Biochim Biophys Acta. 2000. PMID: 11108954 Review. - The Development of Maillard Reaction, and Advanced Glycation End Product (AGE)-Receptor for AGE (RAGE) Signaling Inhibitors as Novel Therapeutic Strategies for Patients with AGE-Related Diseases.
Shen CY, Lu CH, Wu CH, Li KJ, Kuo YM, Hsieh SC, Yu CL. Shen CY, et al. Molecules. 2020 Nov 27;25(23):5591. doi: 10.3390/molecules25235591. Molecules. 2020. PMID: 33261212 Free PMC article. Review.
Cited by
- Progress of RAGE Molecular Imaging in Alzheimer's Disease.
Kong Y, Liu C, Zhou Y, Qi J, Zhang C, Sun B, Wang J, Guan Y. Kong Y, et al. Front Aging Neurosci. 2020 Aug 4;12:227. doi: 10.3389/fnagi.2020.00227. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32848706 Free PMC article. Review. - Advanced Glycation End Products and Inflammation in Type 1 Diabetes Development.
Du C, Whiddett RO, Buckle I, Chen C, Forbes JM, Fotheringham AK. Du C, et al. Cells. 2022 Nov 4;11(21):3503. doi: 10.3390/cells11213503. Cells. 2022. PMID: 36359899 Free PMC article. Review. - Peptide Release after Simulated Infant In Vitro Digestion of Dry Heated Cow's Milk Protein and Transport of Potentially Immunoreactive Peptides across the Caco-2 Cell Monolayer.
Zenker HE, Wichers HJ, Tomassen MMM, Boeren S, De Jong NW, Hettinga KA. Zenker HE, et al. Nutrients. 2020 Aug 18;12(8):2483. doi: 10.3390/nu12082483. Nutrients. 2020. PMID: 32824739 Free PMC article. - Advanced Glycation End-Products (AGEs): Formation, Chemistry, Classification, Receptors, and Diseases Related to AGEs.
Twarda-Clapa A, Olczak A, Białkowska AM, Koziołkiewicz M. Twarda-Clapa A, et al. Cells. 2022 Apr 12;11(8):1312. doi: 10.3390/cells11081312. Cells. 2022. PMID: 35455991 Free PMC article. Review. - Nanopore Sequencing of RAGE Gene Polymorphisms and Their Association with Type 2 Diabetes.
Chaurasiya AH, Khilari AA, Kazi R, Jaiswal MR, Bhoite GM, Padwal MK, Momin AA, Shanmugam D, Kulkarni MJ. Chaurasiya AH, et al. ACS Omega. 2023 Jul 10;8(29):25727-25738. doi: 10.1021/acsomega.3c00297. eCollection 2023 Jul 25. ACS Omega. 2023. PMID: 37521601 Free PMC article.
References
- Atherton E, Fox H, Harkiss D, Sheppard RC. J. Chem. Soc., Chem Commun. 1978:537–539.
- Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. - PubMed
- Brett J, Schmidt AM, Zou YS, Yan SD, Weidman E, Pinsky D, Neeper M, Przysiecki M, shaw A, Migheli A, Stern D. Tissue distribution of the receptor for advanced glycation end products (RAGE): expression in smooth muscle, cardiac myocyte, and neural tissue in addition to vasculature. Am. J. Pathol. 1993;143:1699–1712. - PMC - PubMed
- Brownlee M. Advanced protein glycosylation in diabetes and aging. Annu Rev Med. 1995;46:223–234. - PubMed
- Brownlee M, Vlassara H, Cerami A. Nonenzymatic glycosylation and the pathogenesis of diabetic complications. Ann Intern Med. 1984;101:527–537. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases