The human airway epithelial basal cell transcriptome - PubMed (original) (raw)
The human airway epithelial basal cell transcriptome
Neil R Hackett et al. PLoS One. 2011.
Abstract
Background: The human airway epithelium consists of 4 major cell types: ciliated, secretory, columnar and basal cells. During natural turnover and in response to injury, the airway basal cells function as stem/progenitor cells for the other airway cell types. The objective of this study is to better understand human airway epithelial basal cell biology by defining the gene expression signature of this cell population.
Methodology/principal findings: Bronchial brushing was used to obtain airway epithelium from healthy nonsmokers. Microarrays were used to assess the transcriptome of basal cells purified from the airway epithelium in comparison to the transcriptome of the differentiated airway epithelium. This analysis identified the "human airway basal cell signature" as 1,161 unique genes with >5-fold higher expression level in basal cells compared to differentiated epithelium. The basal cell signature was suppressed when the basal cells differentiated into a ciliated airway epithelium in vitro. The basal cell signature displayed overlap with genes expressed in basal-like cells from other human tissues and with that of murine airway basal cells. Consistent with self-modulation as well as signaling to other airway cell types, the human airway basal cell signature was characterized by genes encoding extracellular matrix components, growth factors and growth factor receptors, including genes related to the EGF and VEGF pathways. Interestingly, while the basal cell signature overlaps that of basal-like cells of other organs, the human airway basal cell signature has features not previously associated with this cell type, including a unique pattern of genes encoding extracellular matrix components, G protein-coupled receptors, neuroactive ligands and receptors, and ion channels.
Conclusion/significance: The human airway epithelial basal cell signature identified in the present study provides novel insights into the molecular phenotype and biology of the stem/progenitor cells of the human airway epithelium.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Characterization of cultured human airway epithelial basal cells.
Large airway epithelial cells were collected by bronchoscopy brushing of healthy nonsmokers and cultured under basal cell-selective conditions for 7 to 8 days until 70% confluent. A–G. Confirmation of basal cell identity and purity by immunohistochemistry of cytospin preparations using cell type-specific markers. A. cytokeratin 5 (basal cells); B. TP63 (basal cells); C. CD151 (basal cells); D. chromagranin A (neuroendocrine cells) E. N-cadherin (mesenchymal cells); F. mucin (MUC) 5AC (secretory cells) and G. β-tubulin IV (ciliated cells). All cells were counterstained with Mayer's hematoxylin. H–K. Differentiation of basal cells on air-liquid interface cultures. H. Immunofluorscent staining for β-tubulin IV, day 0 (DAPI, nucleus). I. β-tubulin IV, day 28 (DAPI, nucleus; red, β-tubulin). J. Scanning electron microscopy at day 28. Scale bar for all panels A–J = 10 µm. K. Immunofluorescent staining of section of 28 day air liquid interface culture for β-tubulin (red) and cytokeratin 5 (green). L. Western analysis of basal and differentiated cell proteins probing equal amounts of extracts of basal cells from nonsmoker (BC-NS), large airway epithelium from a nonsmoker (LAE-NS), and large airway epithelium from a smoker (LAE-S) with antibodies as described in Methods section.
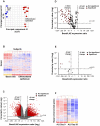
Figure 2. Identification of basal cell-enriched transcripts.
A. Principal component analysis of gene expression of basal cells (n = 5; blue circles) and differentiated airway epithelium samples (n = 12; red circles) using all expressed gene probe sets (n = 39,324) as an input dataset. B. Hierarchical cluster analysis of basal cells (n = 5) compared to complete airway epithelium samples (n = 12) based on the expression of 1,000 randomly chosen probe sets detected in either of groups. Genes expressed above the average are represented in red, below average in blue, and average in white. The genes are represented vertically, and individual samples horizontally. C. Volcano plot comparing the transcriptomes of basal cells (n = 5) and complete airway epithelium (n = 12). In both panels, the y-axis corresponds to the negative log of p value and the x-axis corresponds to the log2-transformed fold-change. Red dots represent significant differentially expressed probe sets (fold-change >5; p value<0.01 with Benjamini-Hochberg correction); grey dots represent nonsignificant gene probe sets. D. Volcano plot assessing the transcriptome of basal cells vs complete large airway epithelium using a list of only ciliogenesis-related genes , . E. Volcano plot assessing the transcriptomes of basal cell vs complete large airway epithelium using a list of only secretory cell-related genes . F. Suppression of the basal cell-enriched transcriptome when basal cells are induced to differentiate into specialized airway cells in air-liquid interface culture. Pure populations of basal cells were plated onto air-liquid interface cultures and RNA was prepared on day 0 and day 28. The gene expression profile was determined and a cluster built using the genes of human airway basal cell-enriched transcriptome.
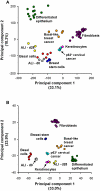
Figure 3. Principal component analysis-based comparison of airway basal cells to other human tissues and cells.
Compared tissues and cell types were all of human origin and included: basal cells (human airway basal cells, red; n = 5); differentiated epithelium (complete large airway epithelium obtained by brushing, green; n = 12); ALI-d0 (basal cells cultured on ALI until confluent, ∼2 days after plating (see Methods), grey; n = 3); ALI-d28 (the same airway basal cells after 28 days of differentiation in air-liquid interface, yellow; n = 3); breast stem cells (from Gene Expression Omnibus GSE15192: CD44+ CD24− stem-like fraction of MCF-10A immortalized breast epithelial cells, dark blue; n = 4), basal-like breast cancer (GSE3744: orange; n = 5); keratinocytes (GSE7216: primary neonatal foreskin epidermal keratinocytes, pink; n = 3); cervical cancer (GSE5993: p63-overexpressing cervical cancer cell line ME180; light blue; n = 3); and fibroblasts (GSE17032: human skin and lung fibroblasts, purple; n = 20). A. Analysis based on the entire transcriptome. B. Analysis based on the 1,161 genes of basal cell signature.
Figure 4. Hierarchical mapping of Gene Ontology (GO) categories enriched in the human basal cell transcriptome.
Basal cell-enriched gene probe sets (n = 1,828) were used to generate a GO tree using GoSurfer software to display “biologic process” categories related to “cellular process” (left branch), “development” (middle branch) and “physiologic process” (right branch). Significantly enriched categories (p<10−10) are represented as red nodes; grey nodes represent mostly closely mapped nonsignificant categories; edges represent “parent-child” relationships of GO terms.
Similar articles
- Endothelial Cell Mediated Promotion of Ciliated Cell Differentiation of Human Airway Basal Cells via Insulin and Insulin-Like Growth Factor 1 Receptor Mediated Signaling.
Gomi K, Tang Y, Arbelaez V, Crystal RG, Walters MS. Gomi K, et al. Stem Cell Rev Rep. 2017 Apr;13(2):309-317. doi: 10.1007/s12015-016-9707-z. Stem Cell Rev Rep. 2017. PMID: 28050756 Free PMC article. - Trefoil factor family 3 peptide promotes human airway epithelial ciliated cell differentiation.
LeSimple P, van Seuningen I, Buisine MP, Copin MC, Hinz M, Hoffmann W, Hajj R, Brody SL, Coraux C, Puchelle E. LeSimple P, et al. Am J Respir Cell Mol Biol. 2007 Mar;36(3):296-303. doi: 10.1165/rcmb.2006-0270OC. Epub 2006 Sep 28. Am J Respir Cell Mol Biol. 2007. PMID: 17008636 - JAG1-Mediated Notch Signaling Regulates Secretory Cell Differentiation of the Human Airway Epithelium.
Gomi K, Staudt MR, Salit J, Kaner RJ, Heldrich J, Rogalski AM, Arbelaez V, Crystal RG, Walters MS. Gomi K, et al. Stem Cell Rev Rep. 2016 Aug;12(4):454-63. doi: 10.1007/s12015-016-9656-6. Stem Cell Rev Rep. 2016. PMID: 27216293 Free PMC article. - Early events in the pathogenesis of chronic obstructive pulmonary disease. Smoking-induced reprogramming of airway epithelial basal progenitor cells.
Shaykhiev R, Crystal RG. Shaykhiev R, et al. Ann Am Thorac Soc. 2014 Dec;11 Suppl 5(Suppl 5):S252-8. doi: 10.1513/AnnalsATS.201402-049AW. Ann Am Thorac Soc. 2014. PMID: 25525728 Free PMC article. Review. - [Structure and function of airway epithelial cells].
Takizawa T. Takizawa T. Nihon Kyobu Shikkan Gakkai Zasshi. 1990 Dec;28(12):1547-56. Nihon Kyobu Shikkan Gakkai Zasshi. 1990. PMID: 2077199 Review. Japanese.
Cited by
- Epithelial Cell Regulation of Allergic Diseases.
Gour N, Lajoie S. Gour N, et al. Curr Allergy Asthma Rep. 2016 Sep;16(9):65. doi: 10.1007/s11882-016-0640-7. Curr Allergy Asthma Rep. 2016. PMID: 27534656 Free PMC article. Review. - An FGFR1-SPRY2 Signaling Axis Limits Basal Cell Proliferation in the Steady-State Airway Epithelium.
Balasooriya GI, Johnson JA, Basson MA, Rawlins EL. Balasooriya GI, et al. Dev Cell. 2016 Apr 4;37(1):85-97. doi: 10.1016/j.devcel.2016.03.001. Dev Cell. 2016. PMID: 27046834 Free PMC article. - Basal-like Cells in the BAL Fluid: An Echo of Regenerative Crisis in Idiopathic Pulmonary Fibrosis Lungs.
Shaykhiev R. Shaykhiev R. Am J Respir Crit Care Med. 2019 Mar 1;199(5):555-557. doi: 10.1164/rccm.201808-1557ED. Am J Respir Crit Care Med. 2019. PMID: 30183332 Free PMC article. No abstract available. - The Kinome of Human Alveolar Type II and Basal Cells, and Its Reprogramming in Lung Cancer.
Leach SM, Finigan J, Vasu VT, Mishra R, Ghosh M, Foster D, Mason R, Kosmider B, Farias Hesson E, Kern JA. Leach SM, et al. Am J Respir Cell Mol Biol. 2019 Oct;61(4):481-491. doi: 10.1165/rcmb.2018-0283OC. Am J Respir Cell Mol Biol. 2019. PMID: 30917006 Free PMC article. - Ontogeny and Biology of Human Small Airway Epithelial Club Cells.
Zuo WL, Shenoy SA, Li S, O'Beirne SL, Strulovici-Barel Y, Leopold PL, Wang G, Staudt MR, Walters MS, Mason C, Kaner RJ, Mezey JG, Crystal RG. Zuo WL, et al. Am J Respir Crit Care Med. 2018 Dec 1;198(11):1375-1388. doi: 10.1164/rccm.201710-2107OC. Am J Respir Crit Care Med. 2018. PMID: 29874100 Free PMC article.
References
- Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8:432–446. - PubMed
- Thompson AB, Robbins RA, Romberger DJ, Sisson JH, Spurzem JR, et al. Immunological functions of the pulmonary epithelium. Eur Respir J. 1995;8:127–149. - PubMed
- Evans MJ, Van Winkle LS, Fanucchi MV, Plopper CG. Cellular and molecular characteristics of basal cells in airway epithelium. Exp Lung Res. 2001;27:401–415. - PubMed
- Rawlins EL, Hogan BL. Epithelial stem cells of the lung: privileged few or opportunities for many? Development. 2006;133:2455–2465. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases