DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs - PubMed (original) (raw)
DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs
Ah-Young So et al. PLoS One. 2011.
Abstract
Epigenetic regulation of gene expression is well known mechanism that regulates cellular senescence of cancer cells. Here we show that inhibition of DNA methyltransferases (DNMTs) with 5-azacytidine (5-AzaC) or with specific small interfering RNA (siRNA) against DNMT1 and 3b induced the cellular senescence of human umbilical cord blood-derived multipotent stem cells (hUCB-MSCs) and increased p16(INK4A) and p21(CIP1/WAF1) expression. DNMT inhibition changed histone marks into the active forms and decreased the methylation of CpG islands in the p16(INK4A) and p21(CIP1/WAF1) promoter regions. Enrichment of EZH2, the key factor that methylates histone H3 lysine 9 and 27 residues, was decreased on the p16(INK4A) and p21(CIP1/WAF1) promoter regions. We found that DNMT inhibition decreased expression levels of Polycomb-group (PcG) proteins and increased expression of microRNAs (miRNAs), which target PcG proteins. Decreased CpG island methylation and increased levels of active histone marks at genomic regions encoding miRNAs were observed after 5-AzaC treatment. Taken together, DNMTs have a critical role in regulating the cellular senescence of hUCB-MSCs through controlling not only the DNA methylation status but also active/inactive histone marks at genomic regions of PcG-targeting miRNAs and p16(INK4A) and p21(CIP1/WAF1) promoter regions.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
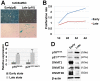
Figure 1. Replicative senescence of hUCB-MSCs.
(a) MSCs undergo replicative senescence upon repeated (more than 15 passages) subculturing in vitro, as shown by SA β-gal staining. (b) Proliferation rates of MSCs in early and late passages were measured by MTT assay. (c–d) The expression of DNMT1, DNMT3A and DNMT3B was down-regulated, whereas p16INK4A was up-regulated during repeated subculture-induced senescence of MSCs, as shown by real-time qPCR (c) and immunoblot analysis (d). * and ** represent statistical significance at the levels of p<0.05 and p<0.01, respectively.
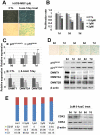
Figure 2. DNMT inhibition induced cellular senescence.
(a) hUCB-MSCs were treated with the DNMT inhibitor 5-AzaC for 7 days. DNMT inhibition by 5-AzaC induced cellular senescence, as shown by SA β-gal staining. (b) After 5-AzaC treatment for 1, 2 and 3 days, an MTT assay was performed. (c–d) 5-AzaC increased p16INK4A and p21WAF1/Cip1 and decreased DNMT1, DNMT3A and DNMT3B, as shown by real-time qPCR analysis (c) and western blot analysis (d). 5-AzaC treatment for 1, 3, 5 and 7 days induced cellular senescence of hUCB-MSCs, as shown by SA β-gal staining (d). (e) After a 2 day treatment with 5-AzaC, FACS analysis was performed, as described in the Materials and Methods section. 5-AzaC treatment induced G1 phase cell cycle arrest in a dose-dependent manner. CDK2 and CDK4 expression levels were confirmed by western blot analysis.
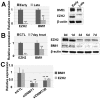
Figure 3. Specific inhibition of DNMT1 and DNMT3b induced cellular senescence.
(a) Specific inhibition of DNMT1 and DNMT3B using siRNA was performed, as described in the Materials and Methods section. The expression levels of DNMT1 and DNMT3B were decreased, as shown by real-time PCR analysis. (b) Specific down-regulation of DNMT1 and DNMT3B caused cellular senescence in MSCs, as shown by SA β-gal staining. (c–d) The expression levels of p16INK4A and p21CIP1/WAF1 were confirmed by real-time qPCR (c) and western blot analysis (d).
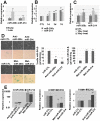
Figure 4. DNMT inhibition modified histone marks, transcriptional enzymes and the CpG island methylation status in the CDKi promoter regions.
(a–b) After treatment with 5-AzaC for 5 days, methyl-specific PCR was performed. (a) Schematic diagrams indicate locations of each primer on CDKi promoter regions. (b) Methyl-specific PCR was performed as described in the Materials and Methods section. M: methyl primer, U: unmethyl primer. (c–f) After treatment with 5-AzaC for 3 days, ChIP analysis was performed using antibodies targeting the indicated protein (AcetylH3, AcetylH4, H3K4Me3, H3K9Me3, H3K27Me3, PolII and EZH2). (c) Schematic diagrams indicate the locations of each primer on genomic DNA. (d–f) Fold enrichment of indicated proteins on the promoters of p16INK4A and p21WAF1/Cip1 were investigated by real-time PCR.
Figure 5. DNMT inhibition decreased PcG expression.
(a) EZH2 and BMI1 expression levels were investigated by real-time qPCR (left) and western blot (right) in early and late passages of hUCB-MSCs. (b) EZH2 and BMI1 expression levels were investigated by real-time qPCR (left) and western blot (right) after the indicated duration treatment of 5-AzaC. (c) Expression levels of BMI1 and EZH2 were investigated by real-time qPCR after DNMT1 and DNMT3B inhibition.
Figure 6. PcG-targeting microRNAs were upregulated after DNMT inhibition.
(a–c) To confirm the expression levels of PcG-targeting microRNAs in early and late passage MSCs and 5-AzaC-treated MSCs, real-time qPCR analysis was performed. Relative expression levels of mature microRNA 200c and 214 in early and late passage (a) and 1, 3 and 7day, 5-AzaC-treated hUCB-MSCs (b) were visualized. Relative expression levels of precursor microRNA 200c and 214 in 1–3 day, 5-AzaC-treated hUCB-MSCs were visualized (c). (d–e) miR200c and miR-214 inhibition and overexpression studies were performed. (d) Overexpression of both miRNAs induced cellular senescence of hUCB-MSCs, as shown by SA β-gal staining. (e) After transfection of anti- and mature-miRNA oligonucleotides, the expression levels of each miRNA and EZH2 and BMI1 were evaluated by real-time qPCR.
Figure 7. DNMT inhibition modified histone marks, transcriptional enzymes as well as the CpG island methylation status in the vicinity of miR-200c and 214 genomic regions.
(a–b) After treatment with 5-AzaC for 5 days, methyl-specific PCR was performed. (a) Schematic diagrams indicate locations of each primer in the vicinity of miR-200c and -214 genomic regions. (b) Methyl-specific PCR was performed as described in the Materials and Methods section. M: methyl primer, U: unmethyl primer. (c–f) After treatment with 5-AzaC for 3 days, ChIP analysis was performed using antibodies targeting to the indicated proteins (AcetylH3, AcetylH4, H3K4Me3, H3K9Me3, H3K27Me3, PolII and EZH2). (c) Schematic diagrams indicate the locations of each primer on genomic DNA. (d–f) Fold enrichment of indicated proteins in the vicinity of miR-200c and -214 genomic regions were investigated by real-time qPCR.
Figure 8. Schematic diagram describing the relationship between miRNAs, PcGs, p16 and p21 and how transcriptional regulation of miRNAs, p16INK4A and p21CIP1/WAF1 occurs in DNMT inhibitor-mediated senescent MSCs.
(a) DNMT inhibition increases p16INK4A and p21CIP1/WAF1 expression directly through DNA demethylation, indirectly through an unknown pathway and, over time, induces cellular senescence. The regulation of miRNAs, which target PcG proteins, is one of the indirect pathways that increase p16INK4A and p21CIP1/WAF1 expression. (b) DNMT inhibition induces CpG island demethylation, increases active histone forms and decreases inactive histone forms in the promoter region of CDK inhibitors and in the proximity of miRNAs in hUCB-MSCs. Pri-miRNA refers to primary-miRNA.
Similar articles
- Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4.
Luis NM, Morey L, Mejetta S, Pascual G, Janich P, Kuebler B, Cozutto L, Roma G, Nascimento E, Frye M, Di Croce L, Benitah SA. Luis NM, et al. Cell Stem Cell. 2011 Sep 2;9(3):233-46. doi: 10.1016/j.stem.2011.07.013. Cell Stem Cell. 2011. PMID: 21885019 - Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1.
Hernández-Muñoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. Hernández-Muñoz I, et al. Mol Cell Biol. 2005 Dec;25(24):11047-58. doi: 10.1128/MCB.25.24.11047-11058.2005. Mol Cell Biol. 2005. PMID: 16314526 Free PMC article. - Expression changes in EZH2, but not in BMI-1, SIRT1, DNMT1 or DNMT3B are associated with DNA methylation changes in prostate cancer.
Hoffmann MJ, Engers R, Florl AR, Otte AP, Muller M, Schulz WA. Hoffmann MJ, et al. Cancer Biol Ther. 2007 Sep;6(9):1403-12. doi: 10.4161/cbt.6.9.4542. Cancer Biol Ther. 2007. PMID: 18637271 - Epigenetic regulation of skin: focus on the Polycomb complex.
Zhang J, Bardot ES, Ezhkova E. Zhang J, et al. Cell Mol Life Sci. 2012 Jul;69(13):2161-2172. doi: 10.1007/s00018-012-0920-x. Cell Mol Life Sci. 2012. PMID: 22314499 Free PMC article. Review. - Epithelial-mesenchymal transition and cancer stemness: the Twist1-Bmi1 connection.
Wu KJ, Yang MH. Wu KJ, et al. Biosci Rep. 2011 Dec;31(6):449-55. doi: 10.1042/BSR20100114. Biosci Rep. 2011. PMID: 21919891 Review.
Cited by
- Neural stem cells exposed to BrdU lose their global DNA methylation and undergo astrocytic differentiation.
Schneider L, d'Adda di Fagagna F. Schneider L, et al. Nucleic Acids Res. 2012 Jul;40(12):5332-42. doi: 10.1093/nar/gks207. Epub 2012 Feb 29. Nucleic Acids Res. 2012. PMID: 22379135 Free PMC article. - Role of DNMTs in the Brain.
Yildiz CB, Zimmer-Bensch G. Yildiz CB, et al. Adv Exp Med Biol. 2022;1389:363-394. doi: 10.1007/978-3-031-11454-0_15. Adv Exp Med Biol. 2022. PMID: 36350518 Review. - Mesenchymal Stem Cell Senescence and Rejuvenation: Current Status and Challenges.
Zhou X, Hong Y, Zhang H, Li X. Zhou X, et al. Front Cell Dev Biol. 2020 Jun 3;8:364. doi: 10.3389/fcell.2020.00364. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32582691 Free PMC article. Review. - Epigenetic Regulation of Bone Marrow Stem Cell Aging: Revealing Epigenetic Signatures associated with Hematopoietic and Mesenchymal Stem Cell Aging.
Cakouros D, Gronthos S. Cakouros D, et al. Aging Dis. 2019 Feb 1;10(1):174-189. doi: 10.14336/AD.2017.1213. eCollection 2019 Feb. Aging Dis. 2019. PMID: 30705777 Free PMC article. Review. - Activation of SAPK/JNK mediated the inhibition and reciprocal interaction of DNA methyltransferase 1 and EZH2 by ursolic acid in human lung cancer cells.
Wu J, Zhao S, Tang Q, Zheng F, Chen Y, Yang L, Yang X, Li L, Wu W, Hann SS. Wu J, et al. J Exp Clin Cancer Res. 2015 Sep 11;34(1):99. doi: 10.1186/s13046-015-0215-9. J Exp Clin Cancer Res. 2015. PMID: 26362062 Free PMC article.
References
- Su Y, Wang X, Zhu WG. [DNA methyltransferases: the role in regulation of gene expression and biological processes]. Yi Chuan. 2009;31:1087–1093. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources