De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state - PubMed (original) (raw)
De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state
Mathias Pawlak et al. Genes Dev. 2011.
Abstract
Induced pluripotent stem cells (iPSCs) are generated from somatic cells by the transduction of defined transcription factors, and this process involves dynamic changes in DNA methylation. While the reprogramming of somatic cells is accompanied by demethylation of pluripotency genes, the functional importance of de novo DNA methylation has not been clarified. Here, using loss-of-function studies, we generated iPSCs from fibroblasts that were deficient in de novo DNA methylation mediated by Dnmt3a and Dnmt3b. These iPSCs reactivated pluripotency genes, underwent self-renewal, and showed restricted developmental potential that was rescued upon reintroduction of Dnmt3a and Dnmt3b. We conclude that de novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells.
Figures
Figure 1.
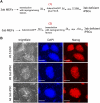
Generation of iPSCs in the absence of de novo DNA methyltransferase activity. (A) Schematic of the two approaches to reprogram conditional knockout MEFs for Dnmt3a and Dnmt3b. MEFs were infected with adenovirus harboring a Cre recombinase and GFP either after or before infection with inducible lentiviruses encoding the reprogramming factors Oct4, Sox2, c-Myc and Klf4. (B) Immunostaining for the pluripotency marker Nanog. Bars, 100μm.
Figure 2.
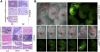
Characterization of 3ab-deficient iPSC lines. (A) Southern blot analysis shows 3ab-deficient iPSC lines obtained through approach 1 (Fig. 1A). Clone #8 is indicated by an asterisk. (B) Southern blot analysis of 3ab-deficient iPSC lines obtained through approach 2 (Fig. 1A). Clones A6 and B2 are indicated by an asterisk. Additionally, 3ab MEFs infected with Adeno-Cre-GFP are shown before the transduction with the reprogramming factors and over the course of several passages (1– 3). Lane 4 displays DNA from a culture dish containing mixed iPSC colonies. Note that the loop-out induced by Cre recombinase was highly efficient. “2lox” represents the conditional alleles and “1lox” represents the deficient alleles of Dnmt3a and Dnmt3b, respectively.
Figure 3.
3ab-deficient iPSCs form teratomas with a restricted developmental potential. (A) Teratomas from #8 and B2 3ab-deficient iPSCs. Reintroduction of functional Dnmt3a and Dnmt3b into 3ab-deficient iPSCs rescues the ability to efficiently form cells from the three germ layers in a teratoma assay. (B) eGFP-marked rescue lines were injected into blastocysts, and resulting chimeric embryos at mid-gestation are shown. The eGFP signal was clearly visible in the majority of embryos, whereas the degree of chimerism varied. Two embryos with developmental defects are shown. The first embryo of the bottom panel did not show any eGFP signal and serves as a reference of background.
Similar articles
- DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development.
Okano M, Bell DW, Haber DA, Li E. Okano M, et al. Cell. 1999 Oct 29;99(3):247-57. doi: 10.1016/s0092-8674(00)81656-6. Cell. 1999. PMID: 10555141 - ICF1-Syndrome-Associated DNMT3B Mutations Prevent De Novo Methylation at a Subset of Imprinted Loci during iPSC Reprogramming.
Verma A, Poondi Krishnan V, Cecere F, D'Angelo E, Lullo V, Strazzullo M, Selig S, Angelini C, Matarazzo MR, Riccio A. Verma A, et al. Biomolecules. 2023 Nov 28;13(12):1717. doi: 10.3390/biom13121717. Biomolecules. 2023. PMID: 38136588 Free PMC article. - Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development.
Yagi M, Kabata M, Tanaka A, Ukai T, Ohta S, Nakabayashi K, Shimizu M, Hata K, Meissner A, Yamamoto T, Yamada Y. Yagi M, et al. Nat Commun. 2020 Jun 24;11(1):3199. doi: 10.1038/s41467-020-16989-w. Nat Commun. 2020. PMID: 32581223 Free PMC article. - The de novo DNA methyltransferase DNMT3A in development and cancer.
Chen BF, Chan WY. Chen BF, et al. Epigenetics. 2014 May;9(5):669-77. doi: 10.4161/epi.28324. Epub 2014 Mar 3. Epigenetics. 2014. PMID: 24589714 Free PMC article. Review. - Using heterokaryons to understand pluripotency and reprogramming.
Piccolo FM, Pereira CF, Cantone I, Brown K, Tsubouchi T, Soza-Ried J, Merkenschlager M, Fisher AG. Piccolo FM, et al. Philos Trans R Soc Lond B Biol Sci. 2011 Aug 12;366(1575):2260-5. doi: 10.1098/rstb.2011.0004. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21727131 Free PMC article. Review.
Cited by
- The use of small molecules in somatic-cell reprogramming.
Federation AJ, Bradner JE, Meissner A. Federation AJ, et al. Trends Cell Biol. 2014 Mar;24(3):179-87. doi: 10.1016/j.tcb.2013.09.011. Epub 2013 Oct 31. Trends Cell Biol. 2014. PMID: 24183602 Free PMC article. Review. - Uhrf1 regulates active transcriptional marks at bivalent domains in pluripotent stem cells through Setd1a.
Kim KY, Tanaka Y, Su J, Cakir B, Xiang Y, Patterson B, Ding J, Jung YW, Kim JH, Hysolli E, Lee H, Dajani R, Kim J, Zhong M, Lee JH, Skalnik D, Lim JM, Sullivan GJ, Wang J, Park IH. Kim KY, et al. Nat Commun. 2018 Jul 3;9(1):2583. doi: 10.1038/s41467-018-04818-0. Nat Commun. 2018. PMID: 29968706 Free PMC article. - The Aging Metabolome-Biomarkers to Hub Metabolites.
Sharma R, Ramanathan A. Sharma R, et al. Proteomics. 2020 Mar;20(5-6):e1800407. doi: 10.1002/pmic.201800407. Proteomics. 2020. PMID: 32068959 Free PMC article. Review. - The mechanistic role of DNA methylation in myeloid leukemogenesis.
Jasielec J, Saloura V, Godley LA. Jasielec J, et al. Leukemia. 2014 Sep;28(9):1765-73. doi: 10.1038/leu.2014.163. Epub 2014 May 20. Leukemia. 2014. PMID: 24913729 Review. - microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming.
Guo X, Liu Q, Wang G, Zhu S, Gao L, Hong W, Chen Y, Wu M, Liu H, Jiang C, Kang J. Guo X, et al. Cell Res. 2013 Jan;23(1):142-56. doi: 10.1038/cr.2012.180. Epub 2012 Dec 25. Cell Res. 2013. PMID: 23266889 Free PMC article.
References
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK 2009. Adult mice generated from induced pluripotent stem cells. Nature 461: 91–94 - PubMed
- Chapman V, Forrester L, Sanford J, Hastie N, Rossant J 1984. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature 307: 284–286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases