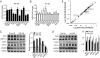miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling - PubMed (original) (raw)
. 2011 May 31;108(22):9232-7.
doi: 10.1073/pnas.1102281108. Epub 2011 May 16.
Leigh Goedeke, Peter Smibert, Cristina M Ramírez, Nikhil P Warrier, Ursula Andreo, Daniel Cirera-Salinas, Katey Rayner, Uthra Suresh, José Carlos Pastor-Pareja, Enric Esplugues, Edward A Fisher, Luiz O F Penalva, Kathryn J Moore, Yajaira Suárez, Eric C Lai, Carlos Fernández-Hernando
Affiliations
- PMID: 21576456
- PMCID: PMC3107310
- DOI: 10.1073/pnas.1102281108
miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling
Alberto Dávalos et al. Proc Natl Acad Sci U S A. 2011.
Abstract
Cellular imbalances of cholesterol and fatty acid metabolism result in pathological processes, including atherosclerosis and metabolic syndrome. Recent work from our group and others has shown that the intronic microRNAs hsa-miR-33a and hsa-miR-33b are located within the sterol regulatory element-binding protein-2 and -1 genes, respectively, and regulate cholesterol homeostasis in concert with their host genes. Here, we show that miR-33a and -b also regulate genes involved in fatty acid metabolism and insulin signaling. miR-33a and -b target key enzymes involved in the regulation of fatty acid oxidation, including carnitine O-octaniltransferase, carnitine palmitoyltransferase 1A, hydroxyacyl-CoA-dehydrogenase, Sirtuin 6 (SIRT6), and AMP kinase subunit-α. Moreover, miR-33a and -b also target the insulin receptor substrate 2, an essential component of the insulin-signaling pathway in the liver. Overexpression of miR-33a and -b reduces both fatty acid oxidation and insulin signaling in hepatic cell lines, whereas inhibition of endogenous miR-33a and -b increases these two metabolic pathways. Together, these data establish that miR-33a and -b regulate pathways controlling three of the risk factors of metabolic syndrome, namely levels of HDL, triglycerides, and insulin signaling, and suggest that inhibitors of miR-33a and -b may be useful in the treatment of this growing health concern.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Posttranscriptional regulation of IRS2, AMPKα, CROT, CPT1a, and HADHB by miR-33b. Quantitative RT-PCR expression profile of selected miR-33 predicted target and other related genes in human hepatic Huh7 cell line (A) after overexpressing miR-33b and (B) after endogenous inhibition of miR-33b by anti–miR-33b. Western blot analysis of HepG2 cells (C) overexpressing or (D) inhibiting endogenous miR-33b. Heat shock protein (HSP)90 bands are the loading control. (E) Specificity of miR-33b on fatty acid metabolism-related genes. qRT-PCR array analysis of fatty acid metabolism-related genes from HepG2 cells transfected with Con miR or miR-33. Data are the mean ± SEM and are representative of more than or equal to three experiments. *P ≤ 0.05.
Fig. 2.
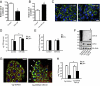
miR-33b regulates human hepatic β-oxidation and lipid homeostasis in Drosophila. (A and B) Relative rate of β-oxidation from Huh7 cells transfected with miR-33 (A) and anti–miR-33b (B). (C) Analysis of neutral lipid accumulation of Huh7 cells transfected with Con-miR or miR-33b and stained with Bodipy (green) and DAPI (blue). (D) Analysis of triglyceride content of Huh7 cells transfected with miR-33b at 0 and 24 h of starvation. (E) Analysis of triglyceride synthesis of Huh7 cells transfected with Con-miR or miR-33 and stimulated or not stimulated with insulin. (F) Northern blot analysis of miR-33, miR-8, and 2S rRNA of transgenic Drosophila overexpressing miR-33 or control transgene in the fat body [genotype: Cg-gal4, upstream activating sequence (UAS)-myrRFP, and UAS-transgene; abbreviated as Cg > transgene]. miR-8 is used as the control. (G) Neutral lipid accumulation in the fat body of Cg > DsRed and Cg > DsRed-miR-33 stained with Bodipy (green), Hoechst 33352 (blue), and the transgene (red). (Scale bar: 30 μm.) (H) Analysis of triglyceride content of transgenic Drosophila overexpressing miR-33 or control transgene in the fat body before and after starvation.
Fig. 3.
miR-33b regulates insulin signaling. (A and B) miR-33b impairs insulin signaling by reducing AKT phosphorylation in hepatic Huh7 cells. (C) In vitro AKT kinase assay of postinsulin-stimulated and immunoprecipitated total AKT from Huh7 cells. GSK-3 fusion protein was used as a substrate and assayed for phosphor-GSK-3α/β (Ser-219). (D and E) IRS2 overexpression rescues Akt phosphorylation in miR-33b–transfected cells after insulin stimulation. (F) miR-33b overexpression reduces 2-deoxyglucose (2-DOG) uptake in Huh7 cells treated with insulin. (G) Quantitative RT-PCR array analysis of insulin signaling related genes from Huh7 cells transfected with Con miR or miR-33. Data are the mean ± SEM and are representative of more than or equal to three experiments. *P ≤ 0.05.
Fig. 4.
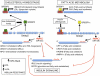
Potential role of SREBPs and miR-33a and -b in metabolic syndrome. In hepatocytes, conditions of low intracellular cholesterol (or statins) induce SREBP-2, leading to increased lipoprotein uptake and endogenous cholesterol biosynthesis. Hyperinsulinemia or insulin resistance induces SREBP-1, leading to increased fatty acid and triglycerides synthesis. The activation of Srebps induces miR-33a and -b expression, leading to decreased HDL cholesterol levels by targeting ABCA1, reduced insulin signaling by targeting IRS2, and reduced cellular β-oxidation by targeting different fatty acid oxidation enzymes. Therapeutic inhibition of miR-33 might result in increased plasma HDL cholesterol levels, reduced VLDL secretion, and increased insulin signaling, thus improving the prognosis of patients with metabolic syndrome.
Similar articles
- MicroRNAs in metabolism and metabolic diseases.
Rottiers V, Najafi-Shoushtari SH, Kristo F, Gurumurthy S, Zhong L, Li Y, Cohen DE, Gerszten RE, Bardeesy N, Mostoslavsky R, Näär AM. Rottiers V, et al. Cold Spring Harb Symp Quant Biol. 2011;76:225-33. doi: 10.1101/sqb.2011.76.011049. Epub 2011 Dec 12. Cold Spring Harb Symp Quant Biol. 2011. PMID: 22156303 Free PMC article. - TXNIP regulates myocardial fatty acid oxidation via miR-33a signaling.
Chen J, Young ME, Chatham JC, Crossman DK, Dell'Italia LJ, Shalev A. Chen J, et al. Am J Physiol Heart Circ Physiol. 2016 Jul 1;311(1):H64-75. doi: 10.1152/ajpheart.00151.2016. Epub 2016 May 3. Am J Physiol Heart Circ Physiol. 2016. PMID: 27199118 Free PMC article. - A regulatory role for microRNA 33* in controlling lipid metabolism gene expression.
Goedeke L, Vales-Lara FM, Fenstermaker M, Cirera-Salinas D, Chamorro-Jorganes A, Ramírez CM, Mattison JA, de Cabo R, Suárez Y, Fernández-Hernando C. Goedeke L, et al. Mol Cell Biol. 2013 Jun;33(11):2339-52. doi: 10.1128/MCB.01714-12. Epub 2013 Apr 1. Mol Cell Biol. 2013. PMID: 23547260 Free PMC article. - MicroRNAs regulating lipid metabolism in atherogenesis.
Rayner KJ, Fernandez-Hernando C, Moore KJ. Rayner KJ, et al. Thromb Haemost. 2012 Apr;107(4):642-7. doi: 10.1160/TH11-10-0694. Epub 2012 Jan 25. Thromb Haemost. 2012. PMID: 22274626 Free PMC article. Review. - MicroRNA modulation of cholesterol homeostasis.
Fernández-Hernando C, Moore KJ. Fernández-Hernando C, et al. Arterioscler Thromb Vasc Biol. 2011 Nov;31(11):2378-82. doi: 10.1161/ATVBAHA.111.226688. Arterioscler Thromb Vasc Biol. 2011. PMID: 22011750 Free PMC article. Review.
Cited by
- Mechanosensitive microRNAs-role in endothelial responses to shear stress and redox state.
Marin T, Gongol B, Chen Z, Woo B, Subramaniam S, Chien S, Shyy JY. Marin T, et al. Free Radic Biol Med. 2013 Sep;64:61-8. doi: 10.1016/j.freeradbiomed.2013.05.034. Epub 2013 May 30. Free Radic Biol Med. 2013. PMID: 23727269 Free PMC article. Review. - Systematic analysis of the regulatory functions of microRNAs in chicken hepatic lipid metabolism.
Li H, Ma Z, Jia L, Li Y, Xu C, Wang T, Han R, Jiang R, Li Z, Sun G, Kang X, Liu X. Li H, et al. Sci Rep. 2016 Aug 18;6:31766. doi: 10.1038/srep31766. Sci Rep. 2016. PMID: 27535581 Free PMC article. - MiR-181a regulates lipid metabolism via IDH1.
Chu B, Wu T, Miao L, Mei Y, Wu M. Chu B, et al. Sci Rep. 2015 Mar 5;5:8801. doi: 10.1038/srep08801. Sci Rep. 2015. PMID: 25739786 Free PMC article. - The Two-Faced Role of SIRT6 in Cancer.
Fiorentino F, Carafa V, Favale G, Altucci L, Mai A, Rotili D. Fiorentino F, et al. Cancers (Basel). 2021 Mar 8;13(5):1156. doi: 10.3390/cancers13051156. Cancers (Basel). 2021. PMID: 33800266 Free PMC article. Review. - A miR-34a-SIRT6 axis in the squamous cell differentiation network.
Lefort K, Brooks Y, Ostano P, Cario-André M, Calpini V, Guinea-Viniegra J, Albinger-Hegyi A, Hoetzenecker W, Kolfschoten I, Wagner EF, Werner S, Dotto GP. Lefort K, et al. EMBO J. 2013 Aug 14;32(16):2248-63. doi: 10.1038/emboj.2013.156. Epub 2013 Jul 16. EMBO J. 2013. PMID: 23860128 Free PMC article.
References
- Alberti KG, Zimmet P, Shaw J. Metabolic syndrome—a new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet Med. 2006;23:469–480. - PubMed
- Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. - PubMed
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. - PubMed
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01GM083300/GM/NIGMS NIH HHS/United States
- R01 HL107953/HL/NHLBI NIH HHS/United States
- R01 HL058541/HL/NHLBI NIH HHS/United States
- R01AG20255/AG/NIA NIH HHS/United States
- P30 HL101270/HL/NHLBI NIH HHS/United States
- R01HL107953/HL/NHLBI NIH HHS/United States
- 1P30HL101270-01/HL/NHLBI NIH HHS/United States
- R01 AG020255/AG/NIA NIH HHS/United States
- R01HL58541/HL/NHLBI NIH HHS/United States
- R01 GM083300/GM/NIGMS NIH HHS/United States
- R01HL16063/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases