Brainstorm: a user-friendly application for MEG/EEG analysis - PubMed (original) (raw)
Brainstorm: a user-friendly application for MEG/EEG analysis
François Tadel et al. Comput Intell Neurosci. 2011.
Abstract
Brainstorm is a collaborative open-source application dedicated to magnetoencephalography (MEG) and electroencephalography (EEG) data visualization and processing, with an emphasis on cortical source estimation techniques and their integration with anatomical magnetic resonance imaging (MRI) data. The primary objective of the software is to connect MEG/EEG neuroscience investigators with both the best-established and cutting-edge methods through a simple and intuitive graphical user interface (GUI).
Figures
Figure 1
General overview of the Brainstorm interface. Considerable effort was made to make the design intuitive and easy to use. The interface includes: (a) a file database that provides direct access to all data (recordings, surfaces, etc.), (b) contextual menus that are available throughout the interface with a right-button click, (c) a batch tool that launches processes (filtering, averaging, statistical tests, etc.) for all files that were drag-and-dropped from the database; (right) multiple displays of information from the database, organized as individual figures and automatically positioned on the screen, and (d) properties of the currently active display.
Figure 2
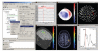
Brainstorm features multiple solutions for the visualization of MEG/EEG recordings.
Figure 3
Interface for reviewing raw recordings and marking events.
Figure 4
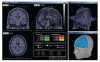
MRI and surface visualization.
Figure 5
Registration of MRI data volumes with corresponding surface meshes.
Figure 6
Brainstorm tool for editing of EEG electrode montages.
Figure 7
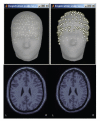
Warping of the MRI volume and corresponding tissue surface envelopes of the Colin27 template brain to fit a set a digitized head points (white dots in upper right corner): initial Colin27 anatomy (left) and warped to the scalp control points of another subject (right). Note how surfaces and MRI volumes are adjusted to the individual data.
Figure 8
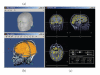
Interactive selection of the best-fitting sphere model parameter for MEG and EEG forward modeling.
Figure 9
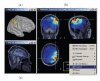
A variety of options for the visualization of estimated sources. (a) 3D rendering of the cortical surface, with control of surface smoothing; (c) 3D orthogonal planes of the MRI volumes; (b) conventional orthogonal views of the MRI volume with overlay of the MEG/EEG source density.
Figure 10
Selection of cortical regions of interest in Brainstorm and extraction of a representative time course of the elementary sources within.
Figure 11
Temporal evolution of elementary dipole sources estimated with the external Xfit software. Data from a right-temporal epileptic spike. This component was implemented in collaboration with Elizabeth Bock, MEG Program, Medical College of Wisconsin.
Figure 12
A variety of display options to visualize time-frequency decompositions using Brainstorm (see text for details).
Figure 13
Graphical interface of the batching tool. (a) selection of the input files by drag-and-drop. (b) creation of an analysis pipeline. (c) example of Matlab script generated automatically.
Figure 14
Example of Brainstorm script.
Figure 15
Student _t_-test between two conditions. (a) selection of the files. (b) selection of the test. (c) options tab for the visualization of statistical maps, including the selection of the thresholding method.
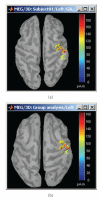
Figure 16
Cortical activations 46 ms after the electric stimulation of the left median nerve on the subject's brain (a) and their projection in the MNI brain (b).
Similar articles
- ElectroMagnetoEncephalography software: overview and integration with other EEG/MEG toolboxes.
Peyk P, De Cesarei A, Junghöfer M. Peyk P, et al. Comput Intell Neurosci. 2011;2011:861705. doi: 10.1155/2011/861705. Epub 2011 Mar 15. Comput Intell Neurosci. 2011. PMID: 21577273 Free PMC article. Review. - Simultaneous EEG and MEG source reconstruction in sparse electromagnetic source imaging.
Ding L, Yuan H. Ding L, et al. Hum Brain Mapp. 2013 Apr;34(4):775-95. doi: 10.1002/hbm.21473. Epub 2011 Nov 18. Hum Brain Mapp. 2013. PMID: 22102512 Free PMC article. - ELAN: a software package for analysis and visualization of MEG, EEG, and LFP signals.
Aguera PE, Jerbi K, Caclin A, Bertrand O. Aguera PE, et al. Comput Intell Neurosci. 2011;2011:158970. doi: 10.1155/2011/158970. Epub 2011 Apr 20. Comput Intell Neurosci. 2011. PMID: 21687568 Free PMC article. - rtMEG: a real-time software interface for magnetoencephalography.
Sudre G, Parkkonen L, Bock E, Baillet S, Wang W, Weber DJ. Sudre G, et al. Comput Intell Neurosci. 2011;2011:327953. doi: 10.1155/2011/327953. Epub 2011 May 17. Comput Intell Neurosci. 2011. PMID: 21687573 Free PMC article. - Scale-free brain activity: past, present, and future.
He BJ. He BJ. Trends Cogn Sci. 2014 Sep;18(9):480-7. doi: 10.1016/j.tics.2014.04.003. Epub 2014 Apr 28. Trends Cogn Sci. 2014. PMID: 24788139 Free PMC article. Review.
Cited by
- Validation of semi-automated anatomically labeled SEEG contacts in a brain atlas for mapping connectivity in focal epilepsy.
Taylor KN, Joshi AA, Hirfanoglu T, Grinenko O, Liu P, Wang X, Gonzalez-Martinez JA, Leahy RM, Mosher JC, Nair DR. Taylor KN, et al. Epilepsia Open. 2021 Sep;6(3):493-503. doi: 10.1002/epi4.12499. Epub 2021 May 15. Epilepsia Open. 2021. PMID: 34033267 Free PMC article. - DUNEuro-A software toolbox for forward modeling in bioelectromagnetism.
Schrader S, Westhoff A, Piastra MC, Miinalainen T, Pursiainen S, Vorwerk J, Brinck H, Wolters CH, Engwer C. Schrader S, et al. PLoS One. 2021 Jun 4;16(6):e0252431. doi: 10.1371/journal.pone.0252431. eCollection 2021. PLoS One. 2021. PMID: 34086715 Free PMC article. - Reduced approach disposition in familial risk for depression: Evidence from time-frequency alpha asymmetries.
Dell'Acqua C, Moretta T, Messerotti Benvenuti S. Dell'Acqua C, et al. PLoS One. 2024 Jul 24;19(7):e0307524. doi: 10.1371/journal.pone.0307524. eCollection 2024. PLoS One. 2024. PMID: 39047003 Free PMC article. - Multivariate sharp-wave ripples in schizophrenia during awake state.
Ohki T, Chao ZC, Takei Y, Kato Y, Sunaga M, Suto T, Tagawa M, Fukuda M. Ohki T, et al. Psychiatry Clin Neurosci. 2024 Sep;78(9):507-516. doi: 10.1111/pcn.13702. Epub 2024 Jun 24. Psychiatry Clin Neurosci. 2024. PMID: 38923051 Free PMC article. - Activity in the lateral occipital cortex between 200 and 300 ms distinguishes between physically identical seen and unseen stimuli.
Liu Y, Paradis AL, Yahia-Cherif L, Tallon-Baudry C. Liu Y, et al. Front Hum Neurosci. 2012 Jul 25;6:211. doi: 10.3389/fnhum.2012.00211. eCollection 2012. Front Hum Neurosci. 2012. PMID: 22848195 Free PMC article.
References
- Baillet S, Mosher JC, Leahy RM. Electromagnetic brain mapping. IEEE Signal Processing Magazine. 2001;18(6):14–30.
- Huang MX, Mosher JC, Leahy RM. A sensor-weighted overlapping-sphere head model and exhaustive head model comparison for MEG. Physics in Medicine and Biology. 1999;44(2):423–440. - PubMed
- Mosher JC, Lewis PS, Leahy RM. Multiple dipole modeling and localization from spatio-temporal MEG data. IEEE Transactions on Biomedical Engineering. 1992;39(5):541–557. - PubMed
- Phillips JW, Leahy RM, Mosher JC. MEG-Based imaging of focal neuronal current sources. IEEE Transactions on Medical Imaging. 1997;16(3):338–348. - PubMed
Publication types
MeSH terms
Grants and funding
- R01-EB009048/EB/NIBIB NIH HHS/United States
- R01 EB000473/EB/NIBIB NIH HHS/United States
- R01-EB002010/EB/NIBIB NIH HHS/United States
- R01-EB000473/EB/NIBIB NIH HHS/United States
- R01 EB009048/EB/NIBIB NIH HHS/United States
- R01 EB002010/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources