Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis - PubMed (original) (raw)
Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis
Florence Jay et al. PLoS Pathog. 2011 May.
Erratum in
- Correction: Misregulation of AUXIN RESPONSE FACTOR 8 Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis.
Jay F, Wang Y, Yu A, Taconnat L, Pelletier S, Colot V, Renou JP, Voinnet O. Jay F, et al. PLoS Pathog. 2016 May 5;12(5):e1005627. doi: 10.1371/journal.ppat.1005627. eCollection 2016 May. PLoS Pathog. 2016. PMID: 27148883 Free PMC article.
Expression of concern in
- Expression of Concern: Misregulation of AUXIN RESPONSE FACTOR 8 Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis.
PLOS Pathogens Editors. PLOS Pathogens Editors. PLoS Pathog. 2015 Oct 9;11(10):e1005234. doi: 10.1371/journal.ppat.1005234. eCollection 2015 Oct. PLoS Pathog. 2015. PMID: 26451730 Free PMC article. No abstract available.
Abstract
In Arabidopsis, micro (mi)RNAs and trans-acting (ta-si)RNAs synthesized directly or indirectly through the DICER-LIKE-1 (DCL1) ribonuclease have roles in patterning and hormonal responses, while DCL2,3,4-dependent small-interfering (si)RNAs are mainly involved in silencing of transposable elements and antiviral defense. Viral suppressors of RNA silencing (VSRs) produced by phytoviruses to counter plant defense may perturb plant developmental programs because of the collision of their inhibitory effects with the regulatory action of endogenous miRNAs and ta-siRNAs. This could explain the similar developmental aberrations displayed by Arabidopsis miRNA/ta-siRNA pathway mutants, including dcl1, and by some VSR-expressing plants. Nonetheless, the molecular bases for these morphological aberrations have remained mysterious, and their contribution to viral disease symptoms/virulence unexplored. The extent of VSR inhibitory actions to other types of endogenous small RNAs remains also unclear. Here, we present an in-depth analysis of transgenic Arabidopsis expressing constitutively HcPro, P19 and P15, three unrelated VSRs. We show that VSR expression has comparable, yet modest effects on known miRNA and ta-siRNA target RNA levels, similar to those observed using an hypomorphic dcl1 mutation. However, by combining results of transcriptome studies with deep-sequencing data from immuno-precipitated small RNAs, additional, novel endogenous targets of miRNA and ta-siRNA were identified, unraveling an unsuspected complexity in the origin and scope-of-action of these molecules. Other stringent analyses pinpointed misregulation of the miR167 target AUXIN RESPONSE FACTOR 8 (ARF8) as a major cause for the developmental aberrations exhibited by VSR transgenic plants, but also for the phenotypes induced during normal viral infection caused by the HcPro-encoding Turnip mosaic virus (TuMV). Neither RNA silencing, its suppression by VSRs, nor the virulence/accumulation of TuMV was altered by mutations in ARF8. These findings have important implications for our understanding of viral disease symptoms and small RNA-directed regulation of plant growth/development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
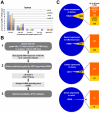
Figure 1. VSRs incur modest yet consistent expression changes to known miRNA and ta-siRNA target transcripts in Arabidopsis.
(A) CATMA gene chip analysis of the ranges in expression changes of known miRNA and ta-siRNA target transcripts that are up-regulated in leaves of transgenic VSR plants and dcl1-9 mutant, as compared to WT plants. Data for other organs are available in Figure S3-S5. nb: number. (B) Diagram summarizing the strategy used in this study for de novo RNA silencing target identification through AGO1-IP small RNA deep sequencing data. (C) Proportions of the total number of genes expressed in each organ analyzed, which were found up-regulated by at least 1.5 fold in at least one VSR transgenic background compared to non-transgenic Arabidopsis. Also indicated is the fraction of up-regulated genes with at least one matching AGO1-IP small RNA read, as assessed by computer-based prediction (see methods).
Figure 2. Using the AGO1-IP read filter in conjunction with VSR microarray data uncovers possibly novel _IR_- and_TAS_-derived siRNA target transcripts with altered accumulation by VSRs.
(A-E) Left panels provide ASRP genome browser views of the small RNA loci of origin. Colored arrows indicate the position and length of the small RNA. Blue, green and red labels indicate 21nt-long, 22nt-long and 24nt-long siRNA species, respectively. A black color signifies small RNAs with length diverging from the above. The right panels depict predicted target sites alongside the small RNA identification number (as in [30]), AGO1-IP read value (underlined in red) and number of loci of origin (hit). The gene identification number of the predicted target is underlined in red. (A–C) Inverted-repeat (IR)-derived siRNAs and their predicted targets, At1g12320 (in leaves; A), At4g08390 (in stems and leaves; B) and At4g28490 (in stems and leaves; C). (D) Predicted secondary structure of the transposon-derived IR6735. (E) A 21nt-long siRNA derived from the TAS3 locus predicted to target the At2g38120 transcript in leaves. For each example, statistically significant up-regulation of gene expression was validated in two independent qRT-PCR analyses of total RNA extracted from the indicated tissues.
Figure 3. Using the AGO1-IP read filter in conjunction with VSR microarray data uncovers possibly novel miRNA* and orphan miRNA target transcripts whose accumulation is altered by VSRs.
As in Figure 3, the upper panels in A–C provide ASRP genome browser views of the small RNA loci of origin. (A-B) Abundantly AGO1-loaded miRNA passenger strands (miR*) for miR159b and miR408, alongside their predicted targets, At3g58780 (stems and leaves; A) and At2g47020 (flowers; B). The blue panels show the predicted stem-loop structures of the corresponding miRNA precursors, in which the miRNA* sequence is boxed in blue. (C) The sulfate transporter At5g13550 transcript is a putative target for the Arabidopsis-specific miR843 in stems. For each example, statistically significant up-regulation of gene expression was validated in two independent qRT-PCR analyses of total RNA extracted from the indicated tissues.
Figure 4. Heterozygous and homozygous arf8 mutant backgrounds respectively attenuate and alleviate the developmental phenotypes incurred by VSRs.
(A) The Venn diagram on the left shows that only a modest number of transcripts are up-regulated in common in leaves of the three VSR transgenics. The table shows that refining the analysis with additional filters based on transcripts up-regulated in dcl1-9 (pale grey) and hen1-1 (dark grey) backgrounds singularizes ARF4 and ARF8, respectively direct targets of miR390 and miR167, as strong candidates for the underlying developmental defects seen in VSR transgenics. (B–C) Strong reduction of leaf and inflorescence defects (inlays) caused by HcPro in F1 progenies of crosses between arf8 mutants and HcPro transgenics carrying the CHS RNAi transgene (B). The Northern blot in (C) shows comparable accumulation of HcPro transcripts in the various backgrounds involved in the crosses. (D–E) same as (B–C) for P15 transgenics with the CHS RNAi background. (F–G) Same as (B–C) for P19 transgenics with the CHS RNAi background. Arrows indicate the presence of slight leaf serration in F1 progeny plants. (H–I) Complete alleviation of developmental defects and sterility of P19 transgenic plants (CHS RNAi background) in the homozygous arf8 mutant background. Northern analysis in (I) confirms comparable P19 levels in the various backgrounds indicated. Plants #1 and #2 where retrieved through independent genotyping in populations of P19 plants with homozygous or heterozygous arf8 mutant genotype. rRNA: ethidium bromide staining of ribosomal RNA provides a control for equal RNA loading.
Figure 5. RNAi and miRNA-mediated gene silencing are not compromised by the arf8 mutation.
(A) Western analysis of DCL1 and AGO1 accumulation in arf8 homozygous seedlings compared to WT seedlings. Negative controls were plants with the dcl1–9 genotype, which accumulate a truncated form of the DCL1 protein, and null ago1–36 mutants. Load: coomassie staining provides a control for equal loading of total proteins. (B) Northern analysis of various endogenous miRNAs in Col-0 or homozygous arf8 mutant seedlings. (C) qRT-PCR analysis of transcript levels from various targets of the miRNAs studied in (B), showing intact miRNA-mediated repression in arf8 mutants as compared to WT plants. (D) RNAi of CHS, diagnosed by a loss-of-seed pigmentation (inlays), remains unaltered in plants with the arf8 -/- genotype. (E) Northern analysis of CHS siRNAs and endogenous miRNA accumulation in VSR transgenics with the heterozygous arf8 mutant background (as depicted in Figure 5B-I). Note the strong decrease in siRNA levels caused by HcPro and P15 as well as the slight shift in electrophoretic migration and enhanced accumulation incurred to miRNAs by P19 and HcPro, respectively. The inlays at the bottom show that RNAi of CHS remains suppressed by the three VSRs in the arf8 mutant background, as diagnosed by the dark-brown seed coloration. (F) qRT-PCR analysis of transcript levels from various targets of the miRNAs studied in (B) in the P19 transgenics carrying the homozygous arf8 mutation (CHS RNAi background), as depicted in Figure 5H. Reference plants used in the analysis were line CHS RNAi and its P19 transgenic derivative (P19 CHS RNAi) with a wild type background. Off-scale values for ARF17 and At4g22470 (a novel small target shown in Figure 4A) are indicated by double-dashed lines. U6: oligonucleotide hybridization of the ubiquitous U6 small nucleolar RNA provides a control for equal RNA loading.
Figure 6. The arf8 mutation does not alter the developmental phenotypes caused by the P6 VSR of Cauliflower mosaic virus but strongly reduces those incurred by_Turnip mosaic virus_ infection.
(A–B) F1 progenies of crosses between arf8–4 mutant and P6 transgenic plants (ecotype Ler) exhibit the typical dwarfism, chlorosis and pointy leaf phenotype incurred by P6 expression. The Northern analysis in (B) shows comparable accumulation of P6 transcripts in the various backgrounds involved in these crosses. (C) The leaf serrations caused by TuMV infection of Col-0 plants (upper panel) are strongly reduced in plants with the arf8–6 -/- mutant background. Note the persistence of chlorosis in both cases. (D) Comparative Northern analysis of TuMV RNA accumulation in Col-0 versus arf8–6 -/- mutant plants. The tracks contain RNA isolated in two independent infections. i: infected; ni: non-infected. rRNA: ethidium bromide staining of ribosomal RNA provides a control for equal RNA loading.
Similar articles
- Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways.
Mlotshwa S, Schauer SE, Smith TH, Mallory AC, Herr JM Jr, Roth B, Merchant DS, Ray A, Bowman LH, Vance VB. Mlotshwa S, et al. Plant Cell. 2005 Nov;17(11):2873-85. doi: 10.1105/tpc.105.036608. Epub 2005 Oct 7. Plant Cell. 2005. PMID: 16214897 Free PMC article. - Developmental Defects Mediated by the P1/HC-Pro Potyviral Silencing Suppressor Are Not Due to Misregulation of AUXIN RESPONSE FACTOR 8.
Mlotshwa S, Pruss GJ, MacArthur JL, Reed JW, Vance V. Mlotshwa S, et al. Plant Physiol. 2016 Nov;172(3):1853-1861. doi: 10.1104/pp.16.01030. Epub 2016 Sep 29. Plant Physiol. 2016. PMID: 27688620 Free PMC article. - Discriminating mutations of HC-Pro of zucchini yellow mosaic virus with differential effects on small RNA pathways involved in viral pathogenicity and symptom development.
Wu HW, Lin SS, Chen KC, Yeh SD, Chua NH. Wu HW, et al. Mol Plant Microbe Interact. 2010 Jan;23(1):17-28. doi: 10.1094/MPMI-23-1-0017. Mol Plant Microbe Interact. 2010. PMID: 19958135 - Structural insights into the arms race between host and virus along RNA silencing pathways in Arabidopsis thaliana.
Guo W, Liew JY, Yuan YA. Guo W, et al. Biol Rev Camb Philos Soc. 2014 May;89(2):337-55. doi: 10.1111/brv.12057. Epub 2013 Sep 4. Biol Rev Camb Philos Soc. 2014. PMID: 24034134 Review. - MicroRNA biogenesis and function in plants.
Chen X. Chen X. FEBS Lett. 2005 Oct 31;579(26):5923-31. doi: 10.1016/j.febslet.2005.07.071. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16144699 Free PMC article. Review.
Cited by
- Plant growth retardation and conserved miRNAs are correlated to Hibiscus chlorotic ringspot virus infection.
Gao R, Wan ZY, Wong SM. Gao R, et al. PLoS One. 2013 Dec 30;8(12):e85476. doi: 10.1371/journal.pone.0085476. eCollection 2013. PLoS One. 2013. PMID: 24386476 Free PMC article. - PAMPs, PRRs, effectors and R-genes associated with citrus-pathogen interactions.
Dalio RJD, Magalhães DM, Rodrigues CM, Arena GD, Oliveira TS, Souza-Neto RR, Picchi SC, Martins PMM, Santos PJC, Maximo HJ, Pacheco IS, De Souza AA, Machado MA. Dalio RJD, et al. Ann Bot. 2017 Mar 1;119(5):749-774. doi: 10.1093/aob/mcw238. Ann Bot. 2017. PMID: 28065920 Free PMC article. Review. - Expression of Concern: Misregulation of AUXIN RESPONSE FACTOR 8 Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis.
PLOS Pathogens Editors. PLOS Pathogens Editors. PLoS Pathog. 2015 Oct 9;11(10):e1005234. doi: 10.1371/journal.ppat.1005234. eCollection 2015 Oct. PLoS Pathog. 2015. PMID: 26451730 Free PMC article. No abstract available. - Citrus psorosis virus 24K protein interacts with citrus miRNA precursors, affects their processing and subsequent miRNA accumulation and target expression.
Reyes CA, Ocolotobiche EE, Marmisollé FE, Robles Luna G, Borniego MB, Bazzini AA, Asurmendi S, García ML. Reyes CA, et al. Mol Plant Pathol. 2016 Apr;17(3):317-29. doi: 10.1111/mpp.12282. Epub 2015 Jul 14. Mol Plant Pathol. 2016. PMID: 26033697 Free PMC article. - Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome.
Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y. Yifhar T, et al. Plant Cell. 2012 Sep;24(9):3575-89. doi: 10.1105/tpc.112.100222. Epub 2012 Sep 21. Plant Cell. 2012. PMID: 23001036 Free PMC article.
References
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. - PubMed
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases