Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions - PubMed (original) (raw)
Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions
Alexander J Ruthenburg et al. Cell. 2011.
Abstract
Little is known about how combinations of histone marks are interpreted at the level of nucleosomes. The second PHD finger of human BPTF is known to specifically recognize histone H3 when methylated on lysine 4 (H3K4me2/3). Here, we examine how additional heterotypic modifications influence BPTF binding. Using peptide surrogates, three acetyllysine ligands are indentified for a PHD-adjacent bromodomain in BPTF via systematic screening and biophysical characterization. Although the bromodomain displays limited discrimination among the three possible acetyllysines at the peptide level, marked selectivity is observed for only one of these sites, H4K16ac, in combination with H3K4me3 at the mononucleosome level. In support, these two histone marks constitute a unique trans-histone modification pattern that unambiguously resides within a single nucleosomal unit in human cells, and this module colocalizes with these marks in the genome. Together, our data call attention to nucleosomal patterning of covalent marks in dictating critical chromatin associations.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
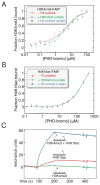
Figure 1. Systematic characterization of preferred acetylated histone ligands for the BPTF bromodomain
(A) Schematic representation of a putative bivalent nucleosomal interaction with the BPTF PHD-bromo module. The known point of contact (Li et al., 2006; Wysocka et al., 2006) is illustrated between the PHD-finger (red) and H3K4me3 (red circle atop green histone tail), while the bromodomain (blue) interacts with an unknown acetylation site (flags on histone tails). (B) A SPOT blot of an array containing all known human core histone acetylation sites on a modified cellulose scaffold probed with GST-tagged bromodomain. A representative SPOT blot (controls and remaining replicates, Figure S1A–C), displaying reproducible staining for H4 peptides (residues 11–25) acetylated on the ε-amines of lysines 16 and 20, respectively (H4K16ac, H4K20ac). The staining of H2BK85ac (red asterisk) appears to be a peptide-HRP interaction and there is no detectable binding between the bromodomain and this peptide in solution (Figure S1E). (C) Peptide pull-down experiments with GST and GST-BPTF bromodomain (GST-bromo) against three peptide series indicated. Full gels with GST controls are available in Figure S1H. (D) Fluorescence polarization anisotropy-based titration of the BPTF bromodomain against each of the indicated peptides (data are represented as mean ± SD). (E) Isothermal titration calorimetry-based binding curves: the indicated H4 peptides are titrated into a solution of BPTF bromodomain (see Figure S1F for _K_d values). See also Figure S1.
Figure 2. Is there allostery or bivalent cooperativity in simultaneous ligand binding by the BPTF PHD-bromodomain?
(A) The possible allosteric cooperative binding at the peptide level is examined by fluorescence polarization anisotropy using 100 nM fluorescein-labelled H3K4me3 peptide, with either no additional peptide, excess unlabeled H4K16ac peptide (500 μM), or excess unlabeled H4 peptide without acetylation (500 μM). (B) The converse experiment with respect to panel A: with unlabeled H3K4me3 in excess (20 μM, ~10-fold above PHD-H3K4me3 _K_d) and fluorescein-H4K16ac (150 nM), protein is titrated and resulting fluorescence polarization anisotropy measured (expressed here as fraction bound). (C) A dsDNA scaffold, selected for rigidity while retaining little predicted bending, was used to covalently install H3K4me3 (H3K4me3[1–8], green), H4K16ac (H4K16ac [12–20], red), or both peptides (blue) at specific positions by disulfide formation with cystamine derivatized convertible dC nucleosides (Figure S2). All of these DNA-protein conjugates were immobilized in different flow channels via a single 3′-biotin linkage to a streptavidin coated surface plasmon resonance chip at low density, untagged PHD-bromo was applied and background binding was subtracted from an empty flow cell. See also Figure S2. (data are represented as mean ± SD)
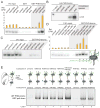
Figure 3. The BPTF PHD-bromo module simultaneously engages two heterotypic trans-histone marks in nucleosomal contexts
(A) GST pull-down of modified nucleosomes with semisynthetic histones produced by EPL. Nucleosomes (unmod, WT recombinant histones; H4K16ac modified; H3K4me3 modified; and dual, H4K16ac and H3K4me3 modified) pulled-down with resin-bound GST or GST-BPTF PHD-bromo module protein are detected by autoradiography after native gel electrophoresis or scintillation counting normalized to indicated % input (Rel. CPM, yellow bars, represent mean ± SD). (B) The reciprocal experiment relative to Figure 3A -- a Western blot of HA-tagged PHD-bromodomain (without GST-tag) retained on streptavidin immobilized mononucleosomes following extensive washing relative to 1% input. (C) The nucleosomes bearing acetylated H4 alone do not display significant binding in the same GST-pull-down experimental format as described in panel A. An additional control nucleosome species with H3T32C and H4R23C histones (unmod-C) serves as an unmodified nucleosome control that retains the cysteine ligation scars. (D) In this experiment, all three H4 acetylation marks that are bound at the peptide-level (Figure 1) are examined in combination with H3K4me3 at the nucleosome level as in panel A. Data are represented as mean ± SD. (E) GST pull-down of hetero-dinucleosomes composed of two mononucleosomes indicated in positions A and B ligated together; each lane is labelled below numerically. The input dinucleosomes for this experiment are presented in Figure S3N. See also Figure S3.
Figure 4. Querying the roles of the PHD finger, the helical linker and the bromodomain in bivalent nucleosome binding via mutagenesis
(A) A ribbon representation of the composite structure of the BPTF PHD-bromodomain module derived from superposition of the previously determined PHD-bromo in complex with H3K4me3 (Li et al., 2006) with the form I BPTF bromodomain H4K16ac complex (Figure 5A). The predicted domain structure of human BPTF shows the position of this module within the whole protein, colored as in Figure 1A and mutations are indicated in grey. Inset panels schematically depict the anticipated consequences of a series of linker helix mutations. (B) Mutations of the PHD-bromo module used to interrogate multivalent binding suggested by the structure assessed by FPA. Upper: a W32E mutation abolishes H3K4me3 binding without disrupting the PHD-finger fold (Li et al., 2006; Pena et al., 2006; Ruthenburg et al., 2007a), while a two amino acid linker helix insertion (labelled +QS), leaves the H3K4me3 binding capacity intact. Lower: a F154A mutation, designed by analogy to previous bromodomain mutagenesis (Dhalluin et al., 1999), abolishes bromodomain binding of H4K16ac; whereas neither the +QS nor the W32E mutants disrupt binding. See Figure S1F for _K_d values. (C) Comparison of the WT GST-PHD-bromodomain (WT) to the series of mutant proteins in the same GST pull-down format as Figure 3. Mutants are labelled WE (W32E), FA (F154A), dbl (W32E + F154A), +QS (a QS insertion after S58 in the bridging helix between the PHD) as depicted in panel A, and 5% input is loaded for comparison (In). (D) Mutations designed to break the α-helix (residues 59–61 all mutated to glycine, TED→ GGG; the same residues mutated to glycines in combination with “SG” or SGGS insertions, TED → GGSGG, and TED→ GGSGGSGG, respectively) are compared to mutations intended to extend the helix and thereby rotate the two domains out of phase (+QS or +AA inserted between and S58 and T59) in the same experimental format as panel C. Mutations that effectively insert two amino acids into the linker helix are indicated in red. See also Figure S4. (data are represented as mean ± SD).
Figure 5. Structural analysis of the BPTF bromodomain peptide complexes
(A) The model derived from crystal form I (bromodomain in blue) with the apical binding site for the H4K16ac peptide (green) rendered in ribbons and sticks. Hydrogen bonds are displayed as yellow dashed lines. (B) Interactions in crystal form II, the bromodomain is colored purple and the bound H4K16ac peptide is depicted in grey. (C) Interaction of the PHD-bromodomain in complex with H4K12ac peptide (peptide, orange; bromodomain, cyan). (D) ITC with BPTF bromodomain mutants designed to disrupt the form I binding mode (V108A and Y147F). Each of these mutations display modest binding deficits comporting with their modest roles in the form I interface. Dissociation constants were outside of the accurately measurable range, so a lower limit of possible _K_d is provided for qualitative comparison. (E) Mutations to disrupt the form II binding interactions while leaving the form II binding mode intact: more substantial mutations, W91A and D101A, produce a more severe loss of affinity. For convenience of comparison, all ΔP scales are identical in scale. (F) Binding conformations of peptides (colored as in previous panels) from each of the structures is compared by Cα-superposition of their respective bromodomains. See also Figure S5.
Figure 6. Bivalent BPTF binding is important for localization of the BPTF PHD-bromodomain and full NURF complex
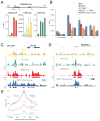
(A) Native ChIP comparison of H3K4me3, H4K12ac, and H4K16ac marks. Four primer sets were employed to interrogate the HOXA9 locus (P1–P3, dark and light grey bars represent HOXA9 exons, and an untranslated region, respectively) and a distal intergenic site [(−) control region]. An average of three real-time PCR replicates of a representative experiment is displayed as a function of % input signal, with error bars reflecting PCR product threshold error amongst the replicates. For simplicity of HOXA9 display, the gene structure annotation represents the Crick strand sense of the genome in this as well as panels B and C. Data are represented as mean ± SD. (B) xChIP of HA-tagged BPTF from HEK293 cell lines that exhibit commensurate expression levels of the ectopic tagged constructs at the HOXA9 locus (with primer sets P2–P4). ChIP signal of tagged WT protein expressing cell lines is compared to discrete cell lines bearing the tagged mutant proteins corresponding to mutations depicted in Figure 4. Data are represented as mean ± SD. (C) ChIP-seq data for the HOXA9 locus, with the three histone modification tracks derived from nChIP-sequencing rendered in yellow, green and red as labelled with input tag counts overlayed in grey on the same scale. On the same abscissal scale and register, the xChIP sequencing counts from the 3xFLAG-tagged BPTF PHD-bromodomain are depicted in blue, with attendant input superimposed in grey. All tag counts are normalized by the factor (2×107/total mapped tags per track) and are unique. Below the continuous tag count graph, MACS peak-called regions relative to input for each sequencing track are depicted in rectangles of the same color. (C) Another example of histone modification patterns and PHD-bromodomain binding, displayed as in the previous panel. (E) Plot of average profile of the PHD-bromodomain at peak regions in the other datasets or intersects thereof. The average PHD signal is contoured on the regions that have MACS called peaks for each individual modification (colored as in 6C); or at loci with called peaks within the same 150 bp window for both H3K4me3 and H4K12ac datasets (purple); or H3K4me3 and H4K12ac datasets (blue). See also Figure S6.
Figure 7. The BPTF PHD-bromodomain preferentially engages a native population of H3K4me3 and H4K16ac doubly-modified nucleosomes
(A) Schematic representation of the experiments: mononucleosome pools were isolated from in nucleo MNase digests and sucrose gradient ultracentrifugation (See Figure S5 for details). (B) Coimmunoprecipitation and western blotting of highly purified mononucleosomes with the well-validated modification and sequence-specific histone antibodies indicated. (C) Recombinantly produced GST-PHD-bromodomain (GST-PB) was used to pull-down native mononucleosomes and the associated material is compared to 5% of the input mononucleosome pool and GST alone for the ability to bind native nucleosomes contingent upon modification patterns. (D) An additional pull-down with the GST tagged bromodomain alone (GST-bromo) was performed alongside GST and GST-PB, and the bound material was probed by Western Blot against the four acetyl marks on the H4 tail for which there are antibodies available. See also Figure S7.
Similar articles
- Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF.
Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Li H, et al. Nature. 2006 Jul 6;442(7098):91-5. doi: 10.1038/nature04802. Epub 2006 May 21. Nature. 2006. PMID: 16728978 Free PMC article. - The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome.
Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J, Musselman CA. Morrison EA, et al. Elife. 2018 Apr 12;7:e31481. doi: 10.7554/eLife.31481. Elife. 2018. PMID: 29648537 Free PMC article. - A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling.
Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. Wysocka J, et al. Nature. 2006 Jul 6;442(7098):86-90. doi: 10.1038/nature04815. Epub 2006 May 21. Nature. 2006. PMID: 16728976 - Opportunity knocks for uncovering the new function of an understudied nucleosome remodeling complex member, the bromodomain PHD finger transcription factor, BPTF.
Zahid H, Olson NM, Pomerantz WCK. Zahid H, et al. Curr Opin Chem Biol. 2021 Aug;63:57-67. doi: 10.1016/j.cbpa.2021.02.003. Epub 2021 Mar 8. Curr Opin Chem Biol. 2021. PMID: 33706239 Free PMC article. Review. - The role of human bromodomains in chromatin biology and gene transcription.
Sanchez R, Zhou MM. Sanchez R, et al. Curr Opin Drug Discov Devel. 2009 Sep;12(5):659-65. Curr Opin Drug Discov Devel. 2009. PMID: 19736624 Free PMC article. Review.
Cited by
- asteRIa enables robust interaction modeling between chromatin modifications and epigenetic readers.
Stadler M, Lukauskas S, Bartke T, Müller CL. Stadler M, et al. Nucleic Acids Res. 2024 Jun 24;52(11):6129-6144. doi: 10.1093/nar/gkae361. Nucleic Acids Res. 2024. PMID: 38752495 Free PMC article. - Exposing the molecular heterogeneity of glycosylated biotherapeutics.
Schachner LF, Mullen C, Phung W, Hinkle JD, Beardsley MI, Bentley T, Day P, Tsai C, Sukumaran S, Baginski T, DiCara D, Agard NJ, Masureel M, Gober J, ElSohly AM, Melani R, Syka JEP, Huguet R, Marty MT, Sandoval W. Schachner LF, et al. Nat Commun. 2024 Apr 16;15(1):3259. doi: 10.1038/s41467-024-47693-8. Nat Commun. 2024. PMID: 38627419 Free PMC article. - Decoding chromatin states by proteomic profiling of nucleosome readers.
Lukauskas S, Tvardovskiy A, Nguyen NV, Stadler M, Faull P, Ravnsborg T, Özdemir Aygenli B, Dornauer S, Flynn H, Lindeboom RGH, Barth TK, Brockers K, Hauck SM, Vermeulen M, Snijders AP, Müller CL, DiMaggio PA, Jensen ON, Schneider R, Bartke T. Lukauskas S, et al. Nature. 2024 Mar;627(8004):671-679. doi: 10.1038/s41586-024-07141-5. Epub 2024 Mar 6. Nature. 2024. PMID: 38448585 Free PMC article. - Guiding the HBO1 complex function through the JADE subunit.
Gaurav N, Kanai A, Lachance C, Cox KL, Liu J, Grzybowski AT, Saksouk N, Klein BJ, Komata Y, Asada S, Ruthenburg AJ, Poirier MG, Côté J, Yokoyama A, Kutateladze TG. Gaurav N, et al. Nat Struct Mol Biol. 2024 Jul;31(7):1039-1049. doi: 10.1038/s41594-024-01245-2. Epub 2024 Mar 6. Nat Struct Mol Biol. 2024. PMID: 38448574 - Beyond the tail: the consequence of context in histone post-translational modification and chromatin research.
Weinzapfel EN, Fedder-Semmes KN, Sun ZW, Keogh MC. Weinzapfel EN, et al. Biochem J. 2024 Feb 21;481(4):219-244. doi: 10.1042/BCJ20230342. Biochem J. 2024. PMID: 38353483 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM040922/GM/NIGMS NIH HHS/United States
- R37 GM053512-30/GM/NIGMS NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- R01 GM040922-26/GM/NIGMS NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R37 GM053512/GM/NIGMS NIH HHS/United States
- R37 GM053512-29/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous