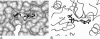Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus - PubMed (original) (raw)
Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus
Vijaykumar Karuppiah et al. J Biol Chem. 2011.
Abstract
Type IV pili are surface-exposed filaments, which extend from a variety of bacterial pathogens and play a major role in pathogenesis, motility, and DNA uptake. Here, we present the crystal structure of a complex between a cytoplasmic component of the type IV pilus biogenesis system from Thermus thermophilus, PilM, in complex with a peptide derived from the cytoplasmic portion of the inner membrane protein PilN. PilM also binds ATP, and its structure is most similar to the actin-like protein FtsA. PilN binds in a narrow channel between the 1A and 1C subdomains in PilM; the binding site is well conserved in other gram-negative bacteria, notably Neisseria meningitidis, Pseudomonas aeruginosa, and Vibrio cholerae. We find no evidence for the catalysis of ATP hydrolysis by PilM; fluorescence data indicate that the protein is likely to be saturated by ATP at physiological concentrations. In addition, binding of the PilN peptide appears to influence the environment of the ATP binding site. This is the first reported structure of a complex between two type IV pilus biogenesis proteins. We propose a model in which PilM binds ATP and then PilN as one of the first steps in the formation of the inner membrane platform of the type IV pilus biogenesis complex.
Figures
FIGURE 1.
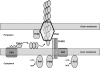
Type IV pilus assembly in T. thermophilus. The pilin monomer contains an N-terminal signal sequence, which is cleaved by the inner membrane prepilin peptidase PilD. The processed pilins assemble into the pilus fiber and emerge from the cell through the PilQ secretin. PilW may assist in the assembly and maturation of the secretin. PilF and PilT are cytoplasmic ATPases, which provide energy for pilus assembly and retraction, respectively. PilC, PilN, and PilO are integral inner membrane proteins. PilM is thought to be part of the multiprotein inner membrane complex, which also includes PilN and PilO.
FIGURE 2.
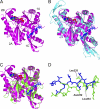
Structure of T. thermophilus PilM and comparison with other bacterial actin-like proteins. A, ribbon plot of PilM (magenta), overlaid with the PilN peptide (blue) and ATP. The four subdomains are labeled according to the original assignment for FtsA (22). B, overlay of PilM and ligands (same colors and orientation as A) with FtsA (cyan; Protein Data Bank code 1E4G). C, overlay of PilM and ligands (same colors and orientation as in A) with EspL (green; Protein Data Bank code 2BH1). D, detail of the PilN peptide (blue) and the C terminus from EpsL (20, 49), with some residues from the latter labeled.
FIGURE 3.
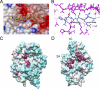
Recognition of PilN by T. thermophilus PilM. A, view of the PilN binding site with a PilM electrostatic surface (blue, positive charge; red, negative charge), generated using CCP4MG (57), and the PilN peptide was superimposed. B, details of the PilM-PilN interaction, with PilM colored magenta and PilN colored blue. Protein-peptide hydrogen bonds and key residues are indicated. C and D, sequence conservation mapped onto the PilM structure using CONSURF (52). Maroon indicates high sequence conservation, white indicates medium sequence conservation, and cyan indicates low sequence conservation, respectively. C, view of the PilN peptide binding site. D, view of the opposite side of the molecule from C, showing the cleft between domains 2A and 1C, which is lined with well conserved residues. Images for C and D were generated using CHIMERA (58).
FIGURE 4.
Alignment of PilN and PilM sequences. A, PilN N terminus sequences. The transmembrane-spanning region, as predicted by the TMPRED program, is indicated by the shaded rectangle. The region of high sequence conservation, recognized by PilM, is boxed. Organisms and sequences used were T. thermophilus (Q72IW7), P. aeruginosa (Q51352), Xanthomonas campestris (Q8P5V4), Legionella pneumophila (Q5WXY0), N. meningitidis (Q4W563), and V. cholerae (C6YKU1). B, PilM sequences, aligned using CLUSTALW (59), with some manual modification. Numbering is for the Thermus sequence. Organisms and sequences used were T. thermophilus (Q5SIJ0), P. aeruginosa (Q51351), X. campestris (Q8P5V3), L. pneumophila (Q5ZX06), N. meningitidis (Q9JY02), and V. cholerae (A1EMB3). Underlined residues had weak or no density and were not included in the PilM/PilN crystal structure.
FIGURE 5.
Recognition of ATP by PilM. A, PilM surface with ATP structure superimposed. B, view of the ATP binding site, with specific interacting residues indicated. Some parts of the structure have been removed for clarity. The figure was generated using CCP4MG (57).
FIGURE 6.
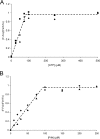
Fluorescence measurement of ATP and PilN binding to PilM. Binding of ATP or PilN was detected through changes in MANT-ATP fluorescence; in each case, ATP or PilN were titrated into a solution of PilM (100 μ
m
) plus varying concentrations of MANT-ATP. _F_o and _F_inf are fluorescence measurements at zero and infinite concentrations of ATP or PilN; F represents fluorescence measurements at all other concentrations. A, ATP titration. MANT-ATP concentrations used were 50 μ
m
(♦), 75 μ
m
(■), and 100 μ
m
(▴). B, PilN titration. MANT-ATP concentrations used were 50 μ
m
(♦) and 100 μ
m
(▴).
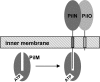
FIGURE 7.
Model for the interaction of PilM with PilN and PilO.
Similar articles
- Structure and assembly of an inner membrane platform for initiation of type IV pilus biogenesis.
Karuppiah V, Collins RF, Thistlethwaite A, Gao Y, Derrick JP. Karuppiah V, et al. Proc Natl Acad Sci U S A. 2013 Nov 26;110(48):E4638-47. doi: 10.1073/pnas.1312313110. Epub 2013 Nov 11. Proc Natl Acad Sci U S A. 2013. PMID: 24218553 Free PMC article. - PilN Binding Modulates the Structure and Binding Partners of the Pseudomonas aeruginosa Type IVa Pilus Protein PilM.
McCallum M, Tammam S, Little DJ, Robinson H, Koo J, Shah M, Calmettes C, Moraes TF, Burrows LL, Howell PL. McCallum M, et al. J Biol Chem. 2016 May 20;291(21):11003-15. doi: 10.1074/jbc.M116.718353. Epub 2016 Mar 28. J Biol Chem. 2016. PMID: 27022027 Free PMC article. - The Type IV Pilus Assembly ATPase PilB of Myxococcus xanthus Interacts with the Inner Membrane Platform Protein PilC and the Nucleotide-binding Protein PilM.
Bischof LF, Friedrich C, Harms A, Søgaard-Andersen L, van der Does C. Bischof LF, et al. J Biol Chem. 2016 Mar 25;291(13):6946-57. doi: 10.1074/jbc.M115.701284. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851283 Free PMC article. - Pulling together with type IV pili.
Nudleman E, Kaiser D. Nudleman E, et al. J Mol Microbiol Biotechnol. 2004;7(1-2):52-62. doi: 10.1159/000077869. J Mol Microbiol Biotechnol. 2004. PMID: 15170403 Review. - Type IV Pili in Thermophilic Bacteria: Mechanisms and Ecological Implications.
Uemura NA, Nakane D. Uemura NA, et al. Biomolecules. 2025 Mar 21;15(4):459. doi: 10.3390/biom15040459. Biomolecules. 2025. PMID: 40305182 Free PMC article. Review.
Cited by
- Type IV pilus proteins form an integrated structure extending from the cytoplasm to the outer membrane.
Li C, Wallace RA, Black WP, Li YZ, Yang Z. Li C, et al. PLoS One. 2013 Jul 26;8(7):e70144. doi: 10.1371/journal.pone.0070144. Print 2013. PLoS One. 2013. PMID: 23922942 Free PMC article. - Architecture of the type IVa pilus machine.
Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Søgaard-Andersen L, Jensen GJ. Chang YW, et al. Science. 2016 Mar 11;351(6278):aad2001. doi: 10.1126/science.aad2001. Epub 2016 Mar 10. Science. 2016. PMID: 26965631 Free PMC article. - The role of core and accessory type IV pilus genes in natural transformation and twitching motility in the bacterium Acinetobacter baylyi.
Leong CG, Bloomfield RA, Boyd CA, Dornbusch AJ, Lieber L, Liu F, Owen A, Slay E, Lang KM, Lostroh CP. Leong CG, et al. PLoS One. 2017 Aug 3;12(8):e0182139. doi: 10.1371/journal.pone.0182139. eCollection 2017. PLoS One. 2017. PMID: 28771515 Free PMC article. - A comprehensive guide to pilus biogenesis in Gram-negative bacteria.
Hospenthal MK, Costa TRD, Waksman G. Hospenthal MK, et al. Nat Rev Microbiol. 2017 May 12;15(6):365-379. doi: 10.1038/nrmicro.2017.40. Nat Rev Microbiol. 2017. PMID: 28496159 Review. - Key role of two terminal domains in the bidirectional polymerization of FtsA protein.
Krupka M, Rivas G, Rico AI, Vicente M. Krupka M, et al. J Biol Chem. 2012 Mar 2;287(10):7756-65. doi: 10.1074/jbc.M111.311563. Epub 2012 Jan 14. J Biol Chem. 2012. PMID: 22247552 Free PMC article.
References
- Nassif X., Pujol C., Morand P., Eugène E. (1999) Mol. Microbiol. 32, 1124–1132 - PubMed
- Ayers M., Sampaleanu L. M., Tammam S., Koo J., Harvey H., Howell P. L., Burrows L. L. (2009) J. Mol. Biol. 394, 128–142 - PubMed
- Morand P. C., Tattevin P., Eugene E., Beretti J. L., Nassif X. (2001) Mol. Microbiol. 40, 846–856 - PubMed
- Davidsen T., Tønjum T. (2006) Nat. Rev. Microbiol. 4, 11–22 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources