Detection of prokaryotic mRNA signifies microbial viability and promotes immunity - PubMed (original) (raw)
Detection of prokaryotic mRNA signifies microbial viability and promotes immunity
Leif E Sander et al. Nature. 2011.
Erratum in
- Nature. 2011 Oct 6;478(7367):136
Abstract
Live vaccines have long been known to trigger far more vigorous immune responses than their killed counterparts. This has been attributed to the ability of live microorganisms to replicate and express specialized virulence factors that facilitate invasion and infection of their hosts. However, protective immunization can often be achieved with a single injection of live, but not dead, attenuated microorganisms stripped of their virulence factors. Pathogen-associated molecular patterns (PAMPs), which are detected by the immune system, are present in both live and killed vaccines, indicating that certain poorly characterized aspects of live microorganisms, not incorporated in dead vaccines, are particularly effective at inducing protective immunity. Here we show that the mammalian innate immune system can directly sense microbial viability through detection of a special class of viability-associated PAMPs (vita-PAMPs). We identify prokaryotic messenger RNA as a vita-PAMP present only in viable bacteria, the recognition of which elicits a unique innate response and a robust adaptive antibody response. Notably, the innate response evoked by viability and prokaryotic mRNA was thus far considered to be reserved for pathogenic bacteria, but we show that even non-pathogenic bacteria in sterile tissues can trigger similar responses, provided that they are alive. Thus, the immune system actively gauges the infectious risk by searching PAMPs for signatures of microbial life and thus infectivity. Detection of vita-PAMPs triggers a state of alert not warranted for dead bacteria. Vaccine formulations that incorporate vita-PAMPs could thus combine the superior protection of live vaccines with the safety of dead vaccines.
Figures
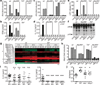
Figure 1. Sensing bacterial viability induces IFN-β and activates the NLRP3 inflammasome in the absence of virulence factors
a. IL-6, TNF-α, b. IFN-β protein, mRNA (at 2h) in BMM stimulated with medium (contr), Lipopolysaccharide (LPS), ThyA— E. coli (EC), and heat-killed EC (HKEC), MOI=20. c. IL-1β (top), and Il1b mRNA (left y-axis), secreted IL-1β (right y-axis) at indicated times (bottom). d, i. Caspase-1 immunoblots at 18h. Pyroptosis by LDH release (e), and FACS (f) at 18h. IL-6 and IL-1β in response to EC, viable or killed by different means (g, BMDC), or viable or HK: EC, attenuated Shigella, Salmonella, and Listeria, or virulent Salmonella SL1344 (h). j. LDH, IL-1β, and IL-6. All responses by BMM and measured at 24h unless indicated otherwise. #, ‘Not detected’. Data represent ≥5 experiments. All bars represent mean ± s.e.m.
Figure 2. The TLR signalling adaptor TRIF controls ‘viability-induced’ responses
Ifnb transcription at 2h (a), IFN-β secretion at 24h (b), Il1b transcription at 2h (c), IL-1β secretion (d), and LDH (e) 24h after phagocytosis of viable or HKEC. f. Caspase-1 immunoblot at 18h. a–f. Data by BMM and represent ≥5 experiments. g. Gene microarray analysis of wt and Trif—/— BMM treated with viable EC for 1, 3 or 6h (three biological replicates #1–3). Heat map of positive regulators/essential components (‘+’) and negative regulators (‘−‘) of inflammasomes. h. Nlrp3 transcription at 1h in BMM. Serum IL-6 and IL-1β 6h after injection of 1×109 viable or 5×109 HKEC (i), and splenic bacterial burdens 72h after injection of 1×108 non-auxotroph EC (j) into wt, Trif—/—, Asc—/— and Nlrp3—/— mice. Each symbol represents one mouse. *; p≤0.05, **; p≤0.01, ***; p≤0.001. n.s., Not statistically significant. #, ‘Not detected’. All bars represent mean ± s.e.m.
Figure 3. Bacterial RNA is a _vita_-PAMP that accesses cytosolic receptors during phagocytosis, and in the absence of virulence factors
a. LPS/Endotoxin, genomic DNA and total RNA in EC before and at indicated times after heat killing. b. Agarose gel electrophoresis of EC total RNA before and after heat killing at 60°C for 60min followed by 4°C incubation for indicated times. c,d. LDH, IL-1β, IFN-β and IL-6 at 24h in response to viable and HKEC, or HKEC with 10µg/ml total EC RNA (HKEC+RNA). # in c,d, ‘Not detected’. a–d, Data by BMM represent ≥5 experiments. e. Representative ratiometric epifluorescence imaging of BMM at 8h with Fdx alone (ctr 8h), Fdx and viable EC (EC 8h) or gentamicin-killed EC (Gent EC) (colour code indicates pH scale). Positive control: Ground silica (silica 1h). f. Quantification of cytosolic Fdx (% of total Fdx/cell). Each dot represents % released Fdx/individual cell. Grey bars represent mean Fdx release. *; p≤0.05, **; p≤0.01, ***; p≤0.001. All bars represent mean ± s.e.m.
Figure 4. Bacterial messenger RNA constitutes an active _vita_-PAMP
a–c and e–g. LDH, IL-1β and IL-6 at 24h. a. Total EC RNA treated with RNAse I and RNAse III, RNAse III alone, or DNAse prior to stimulation of BMDC. b. BMM treated with viable or HKEC, or HKEC with 0.1µg/ml of different bacterial RNA; ribosomal (rRNA), messenger (mRNA), small (sRNA) or eukaryotic RNA (eukRNA). c. BMDC responses. Gro-RNA; in vitro transcribed EC Gro-operon RNA. d. Predicted secondary structure of Gro-RNA. Colour code indicates base pairing probability. e. BMM treated with in vitro transcribed Gro-RNA or Gro dsRNA alone or with HKEC. f. BMDC responses. Era-RNA and DNApol-RNA; in vitro transcribed EC Era-GTPase and DNA-polymerase-III RNA, respectively. g. BMM treated with different doses of unmodified (control), or modified Gro-RNA with HKEC (5’cap, 5’ m7G capping; CIP, calf intestinal phosphatase; 3’poly(A), 3’-polyadenylation). a–g. #, ‘not detected’, all RNA at 10µg/ml except as noted, data represent ≥5 experiments. h. Mice vaccinated and boosted twice with viable EC, HKEC or HKEC with 30µg total purified bacterial RNA (HKEC+RNA) (vaccination regimen in suppl. Fig. 22). Class-specific anti-E. coli antibody serum titers at 25 days. *; p≤0.05, **; p≤0.01, ***; p≤0.001. All bars represent mean ± s.e.m.
Similar articles
- Vita-PAMPs: signatures of microbial viability.
Mourao-Sa D, Roy S, Blander JM. Mourao-Sa D, et al. Adv Exp Med Biol. 2013;785:1-8. doi: 10.1007/978-1-4614-6217-0_1. Adv Exp Med Biol. 2013. PMID: 23456832 Review. - Exploiting vita-PAMPs in vaccines.
Blander JM, Barbet G. Blander JM, et al. Curr Opin Pharmacol. 2018 Aug;41:128-136. doi: 10.1016/j.coph.2018.05.012. Epub 2018 Jun 8. Curr Opin Pharmacol. 2018. PMID: 29890457 Free PMC article. Review. - Measuring Innate Immune Responses to Bacterial Viability.
Moretti J, Vabret N, Blander JM. Moretti J, et al. Methods Mol Biol. 2018;1714:167-190. doi: 10.1007/978-1-4939-7519-8_11. Methods Mol Biol. 2018. PMID: 29177862 - Sensing Microbial Viability through Bacterial RNA Augments T Follicular Helper Cell and Antibody Responses.
Barbet G, Sander LE, Geswell M, Leonardi I, Cerutti A, Iliev I, Blander JM. Barbet G, et al. Immunity. 2018 Mar 20;48(3):584-598.e5. doi: 10.1016/j.immuni.2018.02.015. Epub 2018 Mar 13. Immunity. 2018. PMID: 29548673 Free PMC article. - Bacterial RNA: An Underestimated Stimulus for Innate Immune Responses.
Eigenbrod T, Dalpke AH. Eigenbrod T, et al. J Immunol. 2015 Jul 15;195(2):411-8. doi: 10.4049/jimmunol.1500530. J Immunol. 2015. PMID: 26138638 Review.
Cited by
- Beyond pattern recognition: five immune checkpoints for scaling the microbial threat.
Blander JM, Sander LE. Blander JM, et al. Nat Rev Immunol. 2012 Feb 24;12(3):215-25. doi: 10.1038/nri3167. Nat Rev Immunol. 2012. PMID: 22362354 - Cellular self-defense: how cell-autonomous immunity protects against pathogens.
Randow F, MacMicking JD, James LC. Randow F, et al. Science. 2013 May 10;340(6133):701-6. doi: 10.1126/science.1233028. Science. 2013. PMID: 23661752 Free PMC article. Review. - Human TLR8 is activated upon recognition of Borrelia burgdorferi RNA in the phagosome of human monocytes.
Cervantes JL, La Vake CJ, Weinerman B, Luu S, O'Connell C, Verardi PH, Salazar JC. Cervantes JL, et al. J Leukoc Biol. 2013 Dec;94(6):1231-41. doi: 10.1189/jlb.0413206. Epub 2013 Aug 1. J Leukoc Biol. 2013. PMID: 23906644 Free PMC article. Clinical Trial. - IFN-mediated negative feedback supports bacteria class-specific macrophage inflammatory responses.
Gottschalk RA, Dorrington MG, Dutta B, Krauss KS, Martins AJ, Uderhardt S, Chan W, Tsang JS, Torabi-Parizi P, Fraser ID, Germain RN. Gottschalk RA, et al. Elife. 2019 Aug 6;8:e46836. doi: 10.7554/eLife.46836. Elife. 2019. PMID: 31385572 Free PMC article. - Licensed Bacille Calmette-Guérin (BCG) formulations differ markedly in bacterial viability, RNA content and innate immune activation.
Angelidou A, Conti MG, Diray-Arce J, Benn CS, Shann F, Netea MG, Liu M, Potluri LP, Sanchez-Schmitz G, Husson R, Ozonoff A, Kampmann B, van Haren SD, Levy O. Angelidou A, et al. Vaccine. 2020 Feb 24;38(9):2229-2240. doi: 10.1016/j.vaccine.2019.11.060. Epub 2020 Jan 28. Vaccine. 2020. PMID: 32005538 Free PMC article.
References
- Brockstedt DG, et al. Killed but metabolically active microbes: a new vaccine paradigm for eliciting effector T-cell responses and protective immunity. Nat Med. 2005;11:853–860. - PubMed
- Cheers C, Zhan Y. How do macrophages distinguish the living from the dead? Trends Microbiol. 1996;4:453–455. - PubMed
- Lauvau G, et al. Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science. 2001;294:1735–1739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI064668/AI/NIAID NIH HHS/United States
- T32 GM145304/GM/NIGMS NIH HHS/United States
- R21 AI080959-01A1/AI/NIAID NIH HHS/United States
- R01 AI095245/AI/NIAID NIH HHS/United States
- AI080959A/AI/NIAID NIH HHS/United States
- R21 AI080959/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases