Soluble amyloid precursor protein induces rapid neural differentiation of human embryonic stem cells - PubMed (original) (raw)
Soluble amyloid precursor protein induces rapid neural differentiation of human embryonic stem cells
Kristine K Freude et al. J Biol Chem. 2011.
Abstract
Human embryonic stem cells (hESCs) offer tremendous potential for not only treating neurological disorders but also for their ability to serve as vital reagents to model and investigate human disease. To further our understanding of a key protein involved in Alzheimer disease pathogenesis, we stably overexpressed amyloid precursor protein (APP) in hESCs. Remarkably, we found that APP overexpression in hESCs caused a rapid and robust differentiation of pluripotent stem cells toward a neural fate. Despite maintenance in standard hESC media, up to 80% of cells expressed the neural stem cell marker nestin, and 65% exhibited the more mature neural marker β-3 tubulin within just 5 days of passaging. To elucidate the mechanism underlying the effects of APP on neural differentiation, we examined the proteolysis of APP and performed both gain of function and loss of function experiments. Taken together, our results demonstrate that the N-terminal secreted soluble forms of APP (in particular sAPPβ) robustly drive neural differentiation of hESCs. Our findings not only reveal a novel and intriguing role for APP in neural lineage commitment but also identify a straightforward and rapid approach to generate large numbers of neurons from human embryonic stem cells. These novel APP-hESC lines represent a valuable tool to investigate the potential role of APP in development and neurodegeneration and allow for insights into physiological functions of this protein.
Figures
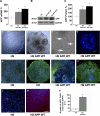
FIGURE 1.
Overexpression of APP markedly alters the morphology of hESC colonies. A, qRT-PCR shows that APP mRNA is significantly elevated in the H9 APP WT and H9 APP Swe clones compared with the parental cell line. APP mRNA was 190 and 240% higher than in untransfected cells. B and C, likewise, APP protein levels were 190 and 230% higher in the APP WT and mutant clones, respectively. D, H9 control colonies display typical densely packed hESC colony morphology, whereas APP-ES clones (H9 APP WT line shown) exhibit large tubular structures and smaller rosette-like morphology (E). F, higher-power image of rosette-like structures in APP-ES clones. G, importantly, karyotype analysis reveals no major chromosomal changes in any of the APP-expressing lines (the H9 APP WT line is shown). H and I, pluripotency markers Oct-4 and SSEA4 confirm the undifferentiated status of H9 control cells. J and K, newly passaged APP-ES clones also exhibit Oct-4 and SSEA4 pluripotent cells (green). However, Oct4 and SSEA4 negative regions (blue only) are also observed within APP-ES colonies as spontaneous differentiation rapidly proceeds. L, low levels of APP (red) are detected in H9 control cells. M, in contrast, APP-ES clones exhibit elevated APP immunoreactivity (red). N, cell counts reveal that 17–20% of H9 control cells express low levels of APP, whereas 80–90% of APP-hESCs express this protein (*, p < 0.05). Scale bar = 300 μm (D, E, I, and K), 180 μm (H and J), and 100 μm (F, L, and M).
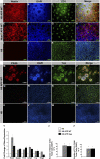
FIGURE 2.
APP overexpression drives rapid and robust neuronal differentiation of H9 and HUES7 hESCs. Despite maintenance in standard hESC media, APP overexpressing H9 (A–D) and HUES7 (E–H) clones spontaneously differentiate, forming large numbers of neural rosettes that express the early neural marker nestin (A, D, E, and H; red). The inner lumens of these rosettes are also immunoreactive for the apical marker ZO-1 (C, D, G, and H; green). I–L, in contrast, control H9 colonies show almost no nestin-positive cells, and ZO-1 labeling is restricted to the cell membrane. M–P, H9 APP WT rosettes also express the neural markers PAX6-positive and TUJ-positive, and uniform patches of more mature PAX6-negative/TUJ-positive NPCs are also observed (Q–T). U–X, control H9 cells, by comparison, express neither PAX6 nor TUJ. Y, qPCR shows that Sox2, nestin, and Tbr2 neural progenitor transcripts are up-regulated in both H9 APP WT and H9 APP Swe clones versus control H9 cells. GFAP, a marker of both neural progenitors and astrocytes, is also elevated in both clones, as is the neural transcript TUJ. The oligodendrocyte transcription factor Olig2 is increased in H9 APP WT clones and down-regulated in H9 APP Swe clones. Z and Z', although almost no H9 cells express nestin or TUJ, these markers are expressed by 83 and 63% of H9 APP WT cells and by 79 and 65% of H9 APP Swe cells, respectively (*, p < 0.05).
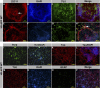
FIGURE 3.
Mature neural markers and neurite outgrowth are rapidly induced by APP overexpression. A–H, H9APP WT clones robustly express APP (red) which is often observed in association with rosette-like structures (A, arrow) or TUJ+ cells (green) (H, arrow). I and J, many H9APP WT NPCs expressed Tbr2, an intermediate basal progenitor cell marker. K and L, furthermore, a subset of cells (≈10%) expresses the mature neural marker tau and (M, N, and P) exhibits extensive neurite outgrowth. O, in contrast, less than 1% of the cells expressed the astrocyte marker GFAP.
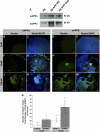
FIGURE 4.
Recombinant sAPPα and β mimic the effects of APP overexpression by inducing rapid neural differentiation of H9 hESCs. A, collected media supernatant from APP-ES cell clones have robustly elevated levels of sAPPα and even more sAPPβ protein. B–E and N, H9 cultures treated with vehicle (0 n
m
) show less than 1% nestin immunoreactivity. F–I, in contrast, addition of 10 n
m
recombinant sAPPα or β protein induces significant differentiation and nestin expression. J–M, treatment of H9 cultures with 100 n
m
sAPPα or β protein induces further neural differentiation. N, quantification reveals that 6% of cells treated with 10 n
m
sAPPα and 22% of those treated with 10 n
m
sAPPβ express nestin. These percentages increased in a dose-dependent manner with 100 n
m
sAPPα yielding 20%, and 100 n
m
sAPPβ yielding 57% nestin-positive cells (*, p < 0.05). Thus, sAPPβ appears to be considerably more potent than sAPPα at inducing neural commitment.
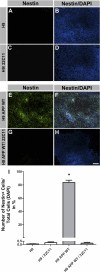
FIGURE 5.
Neural differentiation of APP-hESCs is prevented by treatment with an antibody that binds sAPPα and β. A–D and I, H9 control cultures treated with vehicle or the N-terminal specific antibody, 22C11, remain undifferentiated. E–H and I, in contrast, vehicle-treated H9 APP WT cells rapidly differentiate into nestin-positive NPCs, whereas 22C11 antibody treatment prevents differentiation. Thus, sAPPs mediate neural differentiation of hESCs (*, p < 0.05). Scale bar = 350 μm.
Similar articles
- Kinesin light chain 1 suppression impairs human embryonic stem cell neural differentiation and amyloid precursor protein metabolism.
Killian RL, Flippin JD, Herrera CM, Almenar-Queralt A, Goldstein LS. Killian RL, et al. PLoS One. 2012;7(1):e29755. doi: 10.1371/journal.pone.0029755. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272245 Free PMC article. - Differential processing of amyloid-beta precursor protein directs human embryonic stem cell proliferation and differentiation into neuronal precursor cells.
Porayette P, Gallego MJ, Kaltcheva MM, Bowen RL, Vadakkadath Meethal S, Atwood CS. Porayette P, et al. J Biol Chem. 2009 Aug 28;284(35):23806-17. doi: 10.1074/jbc.M109.026328. Epub 2009 Jun 19. J Biol Chem. 2009. PMID: 19542221 Free PMC article. - In vitro neural differentiation of human embryonic stem cells using a low-density mouse embryonic fibroblast feeder protocol.
Ozolek JA, Jane EP, Esplen JE, Petrosko P, Wehn AK, Erb TM, Mucko SE, Cote LC, Sammak PJ. Ozolek JA, et al. Methods Mol Biol. 2010;584:71-95. doi: 10.1007/978-1-60761-369-5_4. Methods Mol Biol. 2010. PMID: 19907972 - Amyloid precursor proteins, neural differentiation of pluripotent stem cells and its relevance to Alzheimer's disease.
Khandekar N, Lie KH, Sachdev PS, Sidhu KS. Khandekar N, et al. Stem Cells Dev. 2012 May 1;21(7):997-1006. doi: 10.1089/scd.2011.0564. Epub 2012 Jan 18. Stem Cells Dev. 2012. PMID: 22122714 Review. - Mechanism of glial differentiation of neural progenitor cells by amyloid precursor protein.
Sugaya K. Sugaya K. Neurodegener Dis. 2008;5(3-4):170-2. doi: 10.1159/000113693. Epub 2008 Mar 6. Neurodegener Dis. 2008. PMID: 18322381 Review.
Cited by
- Family C G-Protein-Coupled Receptors in Alzheimer's Disease and Therapeutic Implications.
Dal Prà I, Armato U, Chiarini A. Dal Prà I, et al. Front Pharmacol. 2019 Oct 28;10:1282. doi: 10.3389/fphar.2019.01282. eCollection 2019. Front Pharmacol. 2019. PMID: 31719824 Free PMC article. Review. - Caenorhabditis elegans as a model organism to study APP function.
Ewald CY, Li C. Ewald CY, et al. Exp Brain Res. 2012 Apr;217(3-4):397-411. doi: 10.1007/s00221-011-2905-7. Epub 2011 Oct 29. Exp Brain Res. 2012. PMID: 22038715 Free PMC article. Review. - Stem cell models of Alzheimer's disease: progress and challenges.
Arber C, Lovejoy C, Wray S. Arber C, et al. Alzheimers Res Ther. 2017 Jun 13;9(1):42. doi: 10.1186/s13195-017-0268-4. Alzheimers Res Ther. 2017. PMID: 28610595 Free PMC article. Review. - Restoring Soluble Amyloid Precursor Protein α Functions as a Potential Treatment for Alzheimer's Disease.
Habib A, Sawmiller D, Tan J. Habib A, et al. J Neurosci Res. 2017 Apr;95(4):973-991. doi: 10.1002/jnr.23823. Epub 2016 Aug 17. J Neurosci Res. 2017. PMID: 27531392 Free PMC article. Review. - A Unified Hypothesis of Early- and Late-Onset Alzheimer's Disease Pathogenesis.
Atwood CS, Bowen RL. Atwood CS, et al. J Alzheimers Dis. 2015;47(1):33-47. doi: 10.3233/JAD-143210. J Alzheimers Dis. 2015. PMID: 26402752 Free PMC article. Review.
References
- Querfurth H. W., LaFerla F. M. (2010) N. Engl. J. Med. 362, 329–344 - PubMed
- Glabe C. C. (2005) Subcell. Biochem. 38, 167–177 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 AG016573/AG/NIA NIH HHS/United States
- K01 AG029378/AG/NIA NIH HHS/United States
- P30 CA016042/CA/NCI NIH HHS/United States
- P30 DK041301/DK/NIDDK NIH HHS/United States
- P50AG16573/AG/NIA NIH HHS/United States
- K01AG029378/AG/NIA NIH HHS/United States
- R01 AG021982/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources