An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling - PubMed (original) (raw)
An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling
Roger Sciammas et al. Mol Syst Biol. 2011.
Abstract
The B-lymphocyte lineage is a leading system for analyzing gene regulatory networks (GRNs) that orchestrate distinct cell fate transitions. Upon antigen recognition, B cells can diversify their immunoglobulin (Ig) repertoire via somatic hypermutation (SHM) and/or class switch DNA recombination (CSR) before differentiating into antibody-secreting plasma cells. We construct a mathematical model for a GRN underlying this developmental dynamic. The intensity of signaling through the Ig receptor is shown to control the bimodal expression of a pivotal transcription factor, IRF-4, which dictates B cell fate outcomes. Computational modeling coupled with experimental analysis supports a model of 'kinetic control', in which B cell developmental trajectories pass through an obligate transient state of variable duration that promotes diversification of the antibody repertoire by SHM/CSR in direct response to antigens. More generally, this network motif could be used to translate a morphogen gradient into developmental inductive events of varying time, thereby enabling the specification of distinct cell fates.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Figure 1
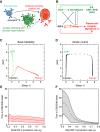
Modeling B cell fate dynamics. (A) Cell fate dynamics that are considered to underlie the generation of effector B cells. Activated B cells can directly differentiate into IgM-secreting plasma cells or undergo CSR/SHM before terminal differentiation. (B) GRN that orchestrates B cell fate dynamics. Genes specific for the CSR/SHM state are shown in green, and those specific for the plasma-cell state are shown in red. Arrows denote activation, and barred lines denote repression. A dotted line indicates that the regulatory interaction may be indirect. In the model, the expression of genes other than Pax5 and Bach2 is assumed to be contingent upon B cell activation; Bcl-6 and Bach2 are treated as a single species. (C, D) Typical trajectories in the [Blimp-1]-[AID] plane of the core deterministic model (Equation (1)) with parameters corresponding to the basic bistability (C) and kinetic-control scenarios (D). Expression coordinates for the indicated components are spaced at equal time intervals. (E, F) Duration of CSR/SHM state as a function of initial IRF-4 production rate. The time spent in the CSR/SHM state decreases with initial IRF-4 production rate (e I) in the kinetic-control scenario (F) but is insensitive to e I in the basic bistability scenario (E). A cell is considered to have entered the CSR/SHM state when its Pax5 concentration is above 3/4 of the peak value of Pax5 (around 1.5) and to have exited when Blimp-1 concentration is greater than a threshold ([Blimp-1]=3). Shading indicates standard deviations arising from noise in the multiscale simulations.
Figure 2
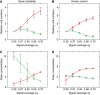
Experimentally testable predictions for B cell fate dynamics. (A, B) Relative numbers of plasma cells (red) and Ig-switched cells (green) predicted by the (A) basic bistability and (B) kinetic-control scenarios as a function of initial IRF-4 production rate (e I). (C, D) Comparison of peak expression levels of Bcl-6/Bach2 (green) and Blimp-1 (red) genes in populations of cells with different initial rates of IRF-4 production in the (C) basic bistability and (D) kinetic-control scenarios. The peak is determined after the averages are calculated for consistency with the RT–PCR measurements.
Figure 3
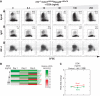
Strength of BCR signaling determines the initial induction levels and subsequent dynamics of IRF-4 expression. (A) The dynamics of IRF-4 expression in response to BCR stimulation. NP-specific B cells were purified from spleens of B1-8i mice and stimulated with NP(12)-Ficoll in the presence of interleukin-2, -4, -5 and soluble CD40L. The expression of IRF-4 was measured over time using flow cytometry of cells labeled with the division-tracking dye CFSE. The two shaded gray bars highlight the IRF-4lo- and IRF-4hi-expressing cell populations. (B) The dynamics of IRF-4 expression in response to differing BCR signaling intensities. Purified NP-reactive B cells were stimulated with serial dilutions of NP(12)-Ficoll (initial concentration 0.01 ng/ml) or NP(109)-Ficoll (0.0001 ng/ml) as described above. IRF-4 expression was analyzed on day 1 (top panels) and day 3 (bottom panels). Data are representative of five experiments.
Figure 4
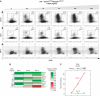
IRF-4 transduces BCR signal strength to control B cell fate. Purified splenic B cells from NP-specific B1-8i heavy chain knock-in mice were stimulated with NP(12)-Ficoll in the presence of interleukin-2, -4, -5 and soluble CD40L. Serial 10-fold dilutions of NP-Ficoll were used with an initial concentration of 0.01 ng/ml. (A) Differentiated cells were analyzed at day 4 and all flow cytometry plots are shown in relation to cell division (CFSE). The frequencies of plasmablasts (Syndecan-1+) and IgG1 class-switched cells are shown as a function of increasing antigen concentration. (B) BCR signal strength dictates B cell fate dynamics by modulating changes in the expression levels of key components of the GRN. RNA was isolated from B cells activated as described above on days 1, 2 and 3. Indicated transcripts were measured by quantitative RT–PCR; transcripts were normalized to Oct1 and used to calculate relative expression. Shown is the average expression from two independent experiments, with results for each gene in turn normalized by its highest expression across all conditions. (C) Peak levels of Bach2 (green) and Blimp-1 (red) expression from (B) as a function of antigen concentration.
Figure 5
Super-induction of IRF-4 regulates B cell fate dynamics in a manner predicted by the modeling. (A) Purified splenic B cells of the indicated genotype (Irf4+/−ColA1 Irf4/Irf4 Rosa26+/M2rtTA) were stimulated with CD40L and IL-4 in the presence of varying concentrations of DOX (6.4–250 ng/ml). The frequencies of IgG1 class-switched cells and of Syndecan-1+ plasmablasts were determined at day 4 of activation, relative to cell division (CFSE dilution). The expression of IRF-4 as a function of DOX was monitored by flow cytometry. The average frequencies from three experiments is shown along with s.e.m. (B, C) Induction of IRF-4 expression by DOX results in changes in the expression levels of key components of the GRN. RNA was isolated and analyzed as described in Figure 4.
Figure 6
Inducible expression of IRF-4 is sufficient to regulate B cell fate in a manner predicted by the modeling. (A) Cell fate of purified splenic B cells of the indicated genotype (Irf4_−/−_ColA1 Irf4/Irf4 Rosa26+/M2rtTA) were enumerated as in Figure 5A. (B, C) Expression levels of key components of the GRN as a function of IRF-4 induction as in Figure 5B and C.
Similar articles
- AID targeting in antibody diversity.
Pavri R, Nussenzweig MC. Pavri R, et al. Adv Immunol. 2011;110:1-26. doi: 10.1016/B978-0-12-387663-8.00005-3. Adv Immunol. 2011. PMID: 21762814 Review. - Immunoglobulin gene diversification.
Maizels N. Maizels N. Annu Rev Genet. 2005;39:23-46. doi: 10.1146/annurev.genet.39.073003.110544. Annu Rev Genet. 2005. PMID: 16285851 Review. - Independent Roles of Switching and Hypermutation in the Development and Persistence of B Lymphocyte Memory.
Gitlin AD, von Boehmer L, Gazumyan A, Shulman Z, Oliveira TY, Nussenzweig MC. Gitlin AD, et al. Immunity. 2016 Apr 19;44(4):769-81. doi: 10.1016/j.immuni.2016.01.011. Epub 2016 Mar 2. Immunity. 2016. PMID: 26944202 Free PMC article. - DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination.
Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, Zarrin AA. Hackney JA, et al. Adv Immunol. 2009;101:163-89. doi: 10.1016/S0065-2776(08)01005-5. Adv Immunol. 2009. PMID: 19231595 Review. - IRF4 Is a Critical Gene in Retinoic Acid-Mediated Plasma Cell Formation and Is Deregulated in Common Variable Immunodeficiency-Derived B Cells.
Indrevær RL, Moskaug JØ, Paur I, Bøhn SK, Jørgensen SF, Blomhoff R, Aukrust P, Fevang B, Blomhoff HK. Indrevær RL, et al. J Immunol. 2015 Sep 15;195(6):2601-11. doi: 10.4049/jimmunol.1500250. Epub 2015 Aug 14. J Immunol. 2015. PMID: 26276871
Cited by
- Diacylglycerol kinase ζ limits B cell antigen receptor-dependent activation of ERK signaling to inhibit early antibody responses.
Wheeler ML, Dong MB, Brink R, Zhong XP, DeFranco AL. Wheeler ML, et al. Sci Signal. 2013 Oct 15;6(297):ra91. doi: 10.1126/scisignal.2004189. Sci Signal. 2013. PMID: 24129701 Free PMC article. - All-or-none suppression of B cell terminal differentiation by environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Zhang Q, Kline DE, Bhattacharya S, Crawford RB, Conolly RB, Thomas RS, Andersen ME, Kaminski NE. Zhang Q, et al. Toxicol Appl Pharmacol. 2013 Apr 1;268(1):17-26. doi: 10.1016/j.taap.2013.01.015. Epub 2013 Jan 26. Toxicol Appl Pharmacol. 2013. PMID: 23357550 Free PMC article. - Single-Cell Expression Variability Implies Cell Function.
Osorio D, Yu X, Zhong Y, Li G, Yu P, Serpedin E, Huang JZ, Cai JJ. Osorio D, et al. Cells. 2019 Dec 19;9(1):14. doi: 10.3390/cells9010014. Cells. 2019. PMID: 31861624 Free PMC article. - The Transcriptional Regulation of Germinal Center Formation.
Song S, Matthias PD. Song S, et al. Front Immunol. 2018 Sep 5;9:2026. doi: 10.3389/fimmu.2018.02026. eCollection 2018. Front Immunol. 2018. PMID: 30233601 Free PMC article. Review. - Transplantation tolerance modifies donor-specific B cell fate to suppress de novo alloreactive B cells.
Khiew SH, Jain D, Chen J, Yang J, Yin D, Young JS, Dent A, Sciammas R, Alegre ML, Chong AS. Khiew SH, et al. J Clin Invest. 2020 Jul 1;130(7):3453-3466. doi: 10.1172/JCI132814. J Clin Invest. 2020. PMID: 32452834 Free PMC article.
References
- Acar M, Becskei A, van Oudenaarden A (2005) Enhancement of cellular memory by reducing stochastic transitions. Nature 435: 228–232 - PubMed
- Alon U (2007) An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman & Hall/CRC
- Aurell E, Brown S, Johanson J, Sneppen K (2002) Stability puzzles in phage λ. Phys Rev E Stat Nonlin Soft Matter Phys 65: 051914. - PubMed
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous