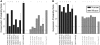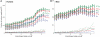Conservation of transcription factor binding events predicts gene expression across species - PubMed (original) (raw)
. 2011 Sep 1;39(16):7092-102.
doi: 10.1093/nar/gkr404. Epub 2011 May 26.
Affiliations
- PMID: 21622661
- PMCID: PMC3167604
- DOI: 10.1093/nar/gkr404
Conservation of transcription factor binding events predicts gene expression across species
Martin Hemberg et al. Nucleic Acids Res. 2011.
Abstract
Recent technological advances have made it possible to determine the genome-wide binding sites of transcription factors (TFs). Comparisons across species have suggested a relatively low degree of evolutionary conservation of experimentally defined TF binding events (TFBEs). Using binding data for six different TFs in hepatocytes and embryonic stem cells from human and mouse, we demonstrate that evolutionary conservation of TFBEs within orthologous proximal promoters is closely linked to function, defined as expression of the target genes. We show that (i) there is a significantly higher degree of conservation of TFBEs when the target gene is expressed in both species; (ii) there is increased conservation of binding events for groups of TFs compared to individual TFs; and (iii) conserved TFBEs have a greater impact on the expression of their target genes than non-conserved ones. These results link conservation of structural elements (TFBEs) to conservation of function (gene expression) and suggest a higher degree of functional conservation than implied by previous studies.
Figures
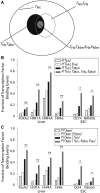
Figure 1.
Genes that are expressed in both human and mouse are more likely to have conserved TFBEs. (A) Schematic illustration of the different conditional gene sets and conditional probability computations for HNF4A (all the numbers and statistics for the other TFs are presented in parts B and C and Table 1). The outer oval (black) represents the total number of genes with a HNF4A TFBE in humans (n = 956; T_Hs). The gray oval represents the subset of genes that show an HNF4A TFBE in humans and which are expressed in the human liver (n = 344; T_Hs, E_Hs). The filled black oval represents the subset of those genes with a HNF4A binding event in human that also show a Hnf4a binding event for the ortholog genes in mouse (n = 307; T_Hs, T_Mm). The filled gray oval represents those genes that have conserved TFBEs and are expressed in both species (n = 106; T_Hs, E_Hs, T_Mm_, E_Mm). (B and C) The _y_-axis indicates the probability of finding a TFBE peak in hepatocytes or ESCs for human (B) or mouse (C) conditional on the presence of TFBE on the other species or on gene expression. The TFBE data are derived from the ChIP–Chip experiments reported in Refs. (11,12,26). The probability of finding a TFBE peak in human, P(_T_Hs), is shown as empty black bars in B [similarly, the probability of finding a TFBE peak in mouse, P(_T_Mm), is shown as empty black bars in C]. The probability of finding a TFBE consistently increases for those genes that are expressed: P(_T_Hs _| E_Hs) in B (empty gray bars, cf. empty black bars) and P(_T_Mm _| E_Mm) in C (empty gray bars, cf. empty black bars). The probability of finding a TFBE in one species given that there is a binding event in the other species is indicated by filled black bars (P(_T_Hs | T_Mm) in B and P(T_Mm| T_Hs) in C). The probability of finding a TFBE in one species given that there is a binding event in the other species and that the gene is expressed in both species is indicated by filled gray bars (P(T_Hs | T_Mm, E_Hs, E_Mm) in B and (P(TMm | T_Hs, E_Hs, E_Mm) in C). *P < 10−3; **P < 10−10 (see ‘Materials and Methods’ section).
Figure 2.
Groups of TFBEs are more likely to be conserved than isolated TFBEs and their targets are more likely to be expressed. (A) The _y_-axis indicates the ratio between the probability of observing a conserved group of two TFBEs and the product of the probabilities of observing conservation of each TFBE. Using the nomenclature defined in the text, the _y_-axis indicates  for human (black) and
for human (black) and  for mouse (gray). Assuming independence, this ratio should take a value of 1 (dashed line). The values for each individual pair as well as for higher-order combinations of TFs are presented in Table 1. The ‘asterisks’ denote significant difference from 1 as assessed by a binomial test (P < 0.05). ‘AVG’ denotes the average ratio for all six TF pairs in the liver data set (the error bars show the standard deviation). (B) Let P(Es | 1Ts, 2Ts, … , nTs) indicate the probability of gene expression in species s (s = Human or Mouse) given the presence of up to n different TFBEs (n = 2 in this figure). If the binding events of different TFs were independent, we would expect that the probability of a gene being expressed given the presence of multiple TFBEs would be the product of the individual probabilities of gene expression given each TFBE. The _y_-axis shows the probability ratio
for mouse (gray). Assuming independence, this ratio should take a value of 1 (dashed line). The values for each individual pair as well as for higher-order combinations of TFs are presented in Table 1. The ‘asterisks’ denote significant difference from 1 as assessed by a binomial test (P < 0.05). ‘AVG’ denotes the average ratio for all six TF pairs in the liver data set (the error bars show the standard deviation). (B) Let P(Es | 1Ts, 2Ts, … , nTs) indicate the probability of gene expression in species s (s = Human or Mouse) given the presence of up to n different TFBEs (n = 2 in this figure). If the binding events of different TFs were independent, we would expect that the probability of a gene being expressed given the presence of multiple TFBEs would be the product of the individual probabilities of gene expression given each TFBE. The _y_-axis shows the probability ratio  for human (black) and mouse (gray). This ratio should take a value of 1 under the null hypothesis of independence (dashed lines). We also considered higher-order combinations of TFs. For n = 3, the mean expression probability ratio was 6.11 for Hs and 3.42 for Mm. For n = 4, the expression probability ratio was 11.57 for Hs and 5.30 for Mm. As emphasized in the text the number of genes with n = 3 or n = 4 TFs was small; therefore this Figure focuses on the results for n = 2. The ‘asterisk’ denotes significant difference from 1 as assessed by a binomial test (P < 0.05). ‘AVG’ denotes the average ratio for all six TF pairs in the liver data set (the error bars show the standard deviation).
for human (black) and mouse (gray). This ratio should take a value of 1 under the null hypothesis of independence (dashed lines). We also considered higher-order combinations of TFs. For n = 3, the mean expression probability ratio was 6.11 for Hs and 3.42 for Mm. For n = 4, the expression probability ratio was 11.57 for Hs and 5.30 for Mm. As emphasized in the text the number of genes with n = 3 or n = 4 TFs was small; therefore this Figure focuses on the results for n = 2. The ‘asterisk’ denotes significant difference from 1 as assessed by a binomial test (P < 0.05). ‘AVG’ denotes the average ratio for all six TF pairs in the liver data set (the error bars show the standard deviation).
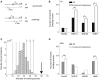
Figure 3.
For most pairs of TFs, rewiring events are more common between genes with conserved expression. (A) Definition of TF rewiring events. We consider a human gene (black line) and its mouse ortholog (gray line). The arrows indicate the transcription start site. The square and circle denote two different TFs. We illustrate TFBE conservation (top: a binding event is found in both species) and rewiring (bottom: for the same gene, one TF is found in one species and a different TF is found in the other species). (B and D) The proportion of rewired binding events is computed as the number of rewiring events divided by the number of rewiring events plus the number of gains and losses [ ]. For a given TF, we added all the rewiring events where that factor was present in human (B) or mouse (D). The filled bars were computed using all genes whereas the empty bars were computed using only those genes that were expressed in both species. *P < 0.01; P < 10−7 (binomial test). (C) We carried out a permutation test whereby the ‘conserved expression’ status was assigned at random (respecting the proportion of genes with conserved expression). Here, we summed the number of rewiring events for all TF pairs. For each permutation we counted the number of rewiring events that occurred between two genes with conserved expression. The histogram shows the distribution of results from 100 000 permutations and the arrow indicates the actual number of rewiring events found in the data. The dashed line denotes the mean of the distribution and the dotted lines denote 1, 2 and 3 SD.
]. For a given TF, we added all the rewiring events where that factor was present in human (B) or mouse (D). The filled bars were computed using all genes whereas the empty bars were computed using only those genes that were expressed in both species. *P < 0.01; P < 10−7 (binomial test). (C) We carried out a permutation test whereby the ‘conserved expression’ status was assigned at random (respecting the proportion of genes with conserved expression). Here, we summed the number of rewiring events for all TF pairs. For each permutation we counted the number of rewiring events that occurred between two genes with conserved expression. The histogram shows the distribution of results from 100 000 permutations and the arrow indicates the actual number of rewiring events found in the data. The dashed line denotes the mean of the distribution and the dotted lines denote 1, 2 and 3 SD.
Figure 4.
Conserved TFBEs have a greater impact on gene expression. The full lines show the square of the Pearson correlation coefficient (_R_2, fraction of variance explained) between the predicted gene-expression levels and the actual gene-expression levels for human (A) and mouse (B). The _x_-axis indicates the gene-expression level enrichment cut-off criterion (see ‘Materials and Methods’ section). For the Hs data, the number of genes included in the analysis decreases from 2322 for an enrichment ratio of 1.2 to 555 for an enrichment ratio of 3.5. For the Mm data, the number of genes included in the analysis decreases from 1806 for an enrichment ratio of 1.2 to 209 for an enrichment ratio of 3.5. The models are described in the text and in the ‘Materials and Methods’ section. To assess whether the _R_2 values could be obtained by chance, we constructed a null hypothesis by randomly shuffling the map between gene expression and TFBE data. The dotted lines show the _R_2 obtained after averaging 1000 shuffle iterations (the color and the dashes correspond to the model with the same solid or dashed line). The error bars represent standard deviations from n = 100 cross-validation steps where 10% of the genes were held out in each iteration.
Similar articles
- CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes.
Hestand MS, van Galen M, Villerius MP, van Ommen GJ, den Dunnen JT, 't Hoen PA. Hestand MS, et al. BMC Bioinformatics. 2008 Nov 26;9:495. doi: 10.1186/1471-2105-9-495. BMC Bioinformatics. 2008. PMID: 19036135 Free PMC article. - Evolutionary conservation of zinc finger transcription factor binding sites in promoters of genes co-expressed with WT1 in prostate cancer.
Eisermann K, Tandon S, Bazarov A, Brett A, Fraizer G, Piontkivska H. Eisermann K, et al. BMC Genomics. 2008 Jul 16;9:337. doi: 10.1186/1471-2164-9-337. BMC Genomics. 2008. PMID: 18631392 Free PMC article. - Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution.
Denas O, Sandstrom R, Cheng Y, Beal K, Herrero J, Hardison RC, Taylor J. Denas O, et al. BMC Genomics. 2015 Feb 14;16(1):87. doi: 10.1186/s12864-015-1245-6. BMC Genomics. 2015. PMID: 25765714 Free PMC article. - A conserved role for transcription factor sumoylation in binding-site selection.
Rosonina E. Rosonina E. Curr Genet. 2019 Dec;65(6):1307-1312. doi: 10.1007/s00294-019-00992-w. Epub 2019 May 15. Curr Genet. 2019. PMID: 31093693 Review. - Chicken ovalbumin upstream promoter-transcription factors and their regulation.
Qiu Y, Krishnan V, Pereira FA, Tsai SY, Tsai MJ. Qiu Y, et al. J Steroid Biochem Mol Biol. 1996 Jan;56(1-6 Spec No):81-5. doi: 10.1016/0960-0760(95)00225-1. J Steroid Biochem Mol Biol. 1996. PMID: 8603050 Review.
Cited by
- Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development.
Göke J, Jung M, Behrens S, Chavez L, O'Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. Göke J, et al. PLoS Comput Biol. 2011 Dec;7(12):e1002304. doi: 10.1371/journal.pcbi.1002304. Epub 2011 Dec 22. PLoS Comput Biol. 2011. PMID: 22215994 Free PMC article. - Uncoupling evolutionary changes in DNA sequence, transcription factor occupancy and enhancer activity.
Khoueiry P, Girardot C, Ciglar L, Peng PC, Gustafson EH, Sinha S, Furlong EE. Khoueiry P, et al. Elife. 2017 Aug 9;6:e28440. doi: 10.7554/eLife.28440. Elife. 2017. PMID: 28792889 Free PMC article. - i-cisTarget: an integrative genomics method for the prediction of regulatory features and cis-regulatory modules.
Herrmann C, Van de Sande B, Potier D, Aerts S. Herrmann C, et al. Nucleic Acids Res. 2012 Aug;40(15):e114. doi: 10.1093/nar/gks543. Epub 2012 Jun 20. Nucleic Acids Res. 2012. PMID: 22718975 Free PMC article. - Cross-species ChIP-seq studies provide insights into regulatory strategies of PPARγ in adipocytes.
Schmidt SF, Jørgensen M, Sandelin A, Mandrup S. Schmidt SF, et al. Transcription. 2012 Jan-Feb;3(1):19-24. doi: 10.4161/trns.3.1.19302. Transcription. 2012. PMID: 22456316 Free PMC article. - Identifying transcription factor complexes and their roles.
Will T, Helms V. Will T, et al. Bioinformatics. 2014 Sep 1;30(17):i415-21. doi: 10.1093/bioinformatics/btu448. Bioinformatics. 2014. PMID: 25161228 Free PMC article.
References
- Arnone M, Davidson E. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. - PubMed
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. - PubMed
- Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. - PubMed
- McGuire AM, Hughes JD, Church GM. Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res. 2000;10:744–757. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous