Molecular structure of amyloid fibrils controls the relationship between fibrillar size and toxicity - PubMed (original) (raw)
Molecular structure of amyloid fibrils controls the relationship between fibrillar size and toxicity
Young Jin Lee et al. PLoS One. 2011.
Abstract
Background: According to the prevailing view, soluble oligomers or small fibrillar fragments are considered to be the most toxic species in prion diseases. To test this hypothesis, two conformationally different amyloid states were produced from the same highly pure recombinant full-length prion protein (rPrP). The cytotoxic potential of intact fibrils and fibrillar fragments generated by sonication from these two states was tested using cultured cells.
Methodology/principal findings: For one amyloid state, fibril fragmentation was found to enhance its cytotoxic potential, whereas for another amyloid state formed within the same amino acid sequence, the fragmented fibrils were found to be substantially less toxic than the intact fibrils. Consistent with the previous studies, the toxic effects were more pronounced for cell cultures expressing normal isoform of the prion protein (PrP(C)) at high levels confirming that cytotoxicity was in part PrP(C)-dependent. Silencing of PrP(C) expression by small hairpin RNAs designed to silence expression of human PrP(C) (shRNA-PrP(C)) diminished the deleterious effects of the two amyloid states to a different extent, suggesting that the role of PrP(C)-mediated and PrP(C)-independent mechanisms depends on the structure of the aggregates.
Conclusions/significance: This work provides a direct illustration that the relationship between an amyloid's physical dimension and its toxic potential is not unidirectional but is controlled by the molecular structure of prion protein (PrP) molecules within aggregated states. Depending on the structure, a decrease in size of amyloid fibrils can either enhance or abolish their cytotoxic effect. Regardless of the molecular structure or size of PrP aggregates, silencing of PrP(C) expression can be exploited to reduce their deleterious effects.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
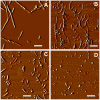
Figure 1. Atomic Force Microscopy imaging of R- and S-fibrils.
Phase AFM images of intact R- and S-fibrils (A and B, respectively), or R- and S-fibrils after fragmentation by ultrasound treatment (C and D respectively). Scale bars = 0.5 µm.
Figure 2. Analysis of PrPC expression in CHO and SKMEL cell lines.
CHO cells before transfection (CHO) or after transfection with pcDNA5/FRT/PrP plasmid (CHO+PrPC), and SKMEL-2 or SKMEL-28 cells (107 cells for each cell line) were lysed and analyzed by Western blotting using mouse anti-PrP 3F4 or mouse anti-β-actin antibody. NBH – 10% normal hamster brain homogenate. β-actin was used as a loading control.
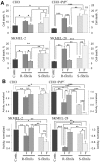
Figure 3. Analysis of cytotoxic potential of intact or fragmented R- and S-fibrils.
Percentage of cell death (A) or activity of mitochondrial dehydrogenases (B) in CHO cells, CHO cells transfected with pcDNA5/FRT/PrP plasmid (CHO+PrPC), SKMEL-2, and SKMEL-28 cells as measured by Trypan Blue (A) or XTT (B) assays, respectively. Cells were seeded at 106cells/cm2 density, cultured for one day prior to administration of rPrP fibrils (1 µM), and then for 24 hours after administration of intact (I) or fragmented (F) R- or S-fibrils. Contr – untreated controls. Each data set represents a mean value ± SD of three independent experiments for both assays. Approximately 500 cells were counted for each data point in each experiment for the Trypan Blue assay. In the XTT assay, XTT activities of untreated controls were set at 100% in each independent experiment. Statistical analyses were performed using Student's t-test. *P<0.05; **P<0.005; ***P<0.0005; NS, non-significant.
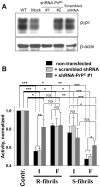
Figure 4. Effect of PrPC silencing by shRNA-PrPC on toxicity of the R- and S-fibrils.
(A) Expression of PrPC in SKMEL-28 cells (WT) and SKMEL-28 cells transfected with lentiviruses encoding shRNA-PrPC#1, shRNA-PrPC#2, or scrambled shRNA, or with empty lentiviral vector (Mock). β-actin was used as a loading control. (B) XTT assay of SKMEL-28 cells transduced with lentiviruses encoding shRNA-PrPC#1 or scrambled shRNA and treated with intact (I) or fragmented (F) R- or S-fibrils (1 µM). Each data set represents a mean value ± SD of three independent experiments. In the XTT assay, XTT activities of untreated controls were set at 100% in each independent experiment. Activities for each set of SKMEL-28 cells (non-transfected, transfected with scrambled shRNA or shRNA-PrPC#1 lentiviruses) were normalized relative to the corresponding controls. Statistical analysis was performed using Student's t-test. *P<0.05; **P<0.01; ***P<0.0005; NS, non-significant.
Similar articles
- Amyloid fibrils of mammalian prion protein are highly toxic to cultured cells and primary neurons.
Novitskaya V, Bocharova OV, Bronstein I, Baskakov IV. Novitskaya V, et al. J Biol Chem. 2006 May 12;281(19):13828-13836. doi: 10.1074/jbc.M511174200. Epub 2006 Mar 22. J Biol Chem. 2006. PMID: 16554307 - Assemblages of prion fragments: novel model systems for understanding amyloid toxicity.
Satheeshkumar KS, Murali J, Jayakumar R. Satheeshkumar KS, et al. J Struct Biol. 2004 Nov;148(2):176-93. doi: 10.1016/j.jsb.2004.05.006. J Struct Biol. 2004. PMID: 15477098 - Formation of soluble oligomers and amyloid fibrils with physical properties of the scrapie isoform of the prion protein from the C-terminal domain of recombinant murine prion protein mPrP-(121-231).
Martins SM, Frosoni DJ, Martinez AM, De Felice FG, Ferreira ST. Martins SM, et al. J Biol Chem. 2006 Sep 8;281(36):26121-8. doi: 10.1074/jbc.M605367200. Epub 2006 Jul 13. J Biol Chem. 2006. PMID: 16844683 - Structure of amyloid oligomers and their mechanisms of toxicities: Targeting amyloid oligomers using novel therapeutic approaches.
Salahuddin P, Fatima MT, Abdelhameed AS, Nusrat S, Khan RH. Salahuddin P, et al. Eur J Med Chem. 2016 May 23;114:41-58. doi: 10.1016/j.ejmech.2016.02.065. Epub 2016 Mar 2. Eur J Med Chem. 2016. PMID: 26974374 Review. - The Structure of Mammalian Prions and Their Aggregates.
Vázquez-Fernández E, Young HS, Requena JR, Wille H. Vázquez-Fernández E, et al. Int Rev Cell Mol Biol. 2017;329:277-301. doi: 10.1016/bs.ircmb.2016.08.013. Epub 2016 Oct 22. Int Rev Cell Mol Biol. 2017. PMID: 28109330 Review.
Cited by
- Inhibition of Insulin Amyloid Fibrillation by a Novel Amphipathic Heptapeptide: MECHANISTIC DETAILS STUDIED BY SPECTROSCOPY IN COMBINATION WITH MICROSCOPY.
Ratha BN, Ghosh A, Brender JR, Gayen N, Ilyas H, Neeraja C, Das KP, Mandal AK, Bhunia A. Ratha BN, et al. J Biol Chem. 2016 Nov 4;291(45):23545-23556. doi: 10.1074/jbc.M116.742460. Epub 2016 Sep 27. J Biol Chem. 2016. PMID: 27679488 Free PMC article. - Analysis of Toxic Amyloid Fibril Interactions at Natively Derived Membranes by Ellipsometry.
Smith RA, Nabok A, Blakeman BJ, Xue WF, Abell B, Smith DP. Smith RA, et al. PLoS One. 2015 Jul 14;10(7):e0132309. doi: 10.1371/journal.pone.0132309. eCollection 2015. PLoS One. 2015. PMID: 26172440 Free PMC article. - β2-microglobulin amyloid fibrils are nanoparticles that disrupt lysosomal membrane protein trafficking and inhibit protein degradation by lysosomes.
Jakhria T, Hellewell AL, Porter MY, Jackson MP, Tipping KW, Xue WF, Radford SE, Hewitt EW. Jakhria T, et al. J Biol Chem. 2014 Dec 26;289(52):35781-94. doi: 10.1074/jbc.M114.586222. Epub 2014 Nov 5. J Biol Chem. 2014. PMID: 25378395 Free PMC article. - Cell Damage in Light Chain Amyloidosis: FIBRIL INTERNALIZATION, TOXICITY AND CELL-MEDIATED SEEDING.
Marin-Argany M, Lin Y, Misra P, Williams A, Wall JS, Howell KG, Elsbernd LR, McClure M, Ramirez-Alvarado M. Marin-Argany M, et al. J Biol Chem. 2016 Sep 16;291(38):19813-25. doi: 10.1074/jbc.M116.736736. Epub 2016 Jul 26. J Biol Chem. 2016. PMID: 27462073 Free PMC article. - Strain-dependent profile of misfolded prion protein aggregates.
Morales R, Hu PP, Duran-Aniotz C, Moda F, Diaz-Espinoza R, Chen B, Bravo-Alegria J, Makarava N, Baskakov IV, Soto C. Morales R, et al. Sci Rep. 2016 Feb 15;6:20526. doi: 10.1038/srep20526. Sci Rep. 2016. PMID: 26877167 Free PMC article.
References
- Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. - PubMed
- Prusiner SB. Prion diseases and the BSE crisis. Science. 1997;278:245–251. - PubMed
- Caughey B, Lansbury PT. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu Rev Neurosci. 2003;26:267–298. - PubMed
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxiciy. Nat Rev Mol Cell Biol. 2007;8:552–561. - PubMed
- Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J Neurosci Res. 2002;69:567–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials