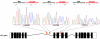Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice - PubMed (original) (raw)
Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice
Louise van der Weyden et al. Blood. 2011.
Abstract
The t(12;21) translocation that generates the ETV6-RUNX1 (TEL-AML1) fusion gene, is the most common chromosomal rearrangement in childhood cancer and is exclusively associated with B-cell precursor acute lymphoblastic leukemia (BCP-ALL). The translocation arises in utero and is necessary but insufficient for the development of leukemia. Single-nucleotide polymorphism array analysis of ETV6-RUNX1 patient samples has identified multiple additional genetic alterations; however, the role of these lesions in leukemogenesis remains undetermined. Moreover, murine models of ETV6-RUNX1 ALL that faithfully recapitulate the human disease are lacking. To identify novel genes that cooperate with ETV6-RUNX1 in leukemogenesis, we generated a mouse model that uses the endogenous Etv6 locus to coexpress the Etv6-RUNX1 fusion and Sleeping Beauty transposase. An insertional mutagenesis screen was performed by intercrossing these mice with those carrying a Sleeping Beauty transposon array. In contrast to previous models, a substantial proportion (20%) of the offspring developed BCP-ALL. Isolation of the transposon insertion sites identified genes known to be associated with BCP-ALL, including Ebf1 and Epor, in addition to other novel candidates. This is the first mouse model of ETV6-RUNX1 to develop BCP-ALL and provides important insight into the cooperating genetic alterations in ETV6-RUNX1 leukemia.
Figures
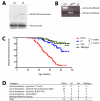
Figure 1. Generation of the Etv6-RUNX1 transposase knockin allele (Etv6+/RUNX1)
A. The endogenous Etv6 locus was targeted to introduce a splice acceptor (SA), exons 1-6 of human RUNX1, an IRES followed by a hyperactive variant of the Sleeping Beauty transposase (HSB5), and an _FRT_-flanked _PGK_-PurodTK drug selection marker between exons 5 and 6. The entire cassette was flanked by Lox66 and Lox71 sites for potential Cre-mediated inactivation of the Etv6-RUNX1 fusion gene. The PurodTK drug marker was removed by breeding mice to a Flpe deleter strain prior to tumor watch experiments. Probe A (5′) and Probe B (3′) are shown. S, _Stu_I. B. Southern blot analysis on _Stu_I digested tail DNA confirmed germline transmission and homologous recombination of the 5′ and 3′ targeting vector arms. C. RT-PCR showing expression of Etv6-RUNX1 fusion transcripts in bone marrow from Etv6+/RUNX1 mice, using primers located as indicated by the arrows on the schematic of the Etv6 locus. Lower panel shows a sequence trace showing splicing of the Etv6 and RUNX1 transcripts to produce an in-frame fusion transcript. D. qPCR of Etv6 and Etv6-RUNX1 fusion transcripts (using primers as mentioned above). RNA was extracted from bone marrow, spleen and thymus. Relative Etv6 gene expression in wildtype (blue) and Etv6+/RUNX1 tissues (red), and relative Etv6-RUNX1 fusion transcript expression in Etv6+/RUNX1 tissues (green) is shown. The data was collected from the analysis of 5 littermate mice of each genotype (±SD). ** P<0.005, * P<0.05.
Figure 2. Activation of transposon mutagenesis in Etv6+/RUNX1 mice
A. Western blot showing expression of the Sleeping Beauty transposase in the bone marrow of Etv6+/RUNX1 (ER) mice, but not mice carrying the T2Onc transposon (Onc). B. ‘Excision’ PCR showing mobilization of the transposon in bone marrow from Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice, as evidenced by the 200 bp ‘excision’ band (instead of the 2 kb band seen in mice carrying the unmobilized transposon array, i.e., Etv6+/+; T2Onc+/Tg (Onc) mice). C. Kaplan-Meier curves showing the tumor latency of ‘jumping’ Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice and ‘non-jumping’ control mice (ER, Onc and wildtype). D. Categorization of the malignancies developed by mice shown in the Kaplan-Meier curve according to the Bethesda criteria for lymphoid and non-lymphoid murine malignancies.-
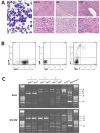
Figure 3. Etv6+/RUNX1 mice with ‘second hits’ develop a pre-B acute lymphoblastic leukemia recapitulating features of the human disease
A. Peripheral blood (PB; ×2000 magnification), spleen, bone marrow and liver (×400 magnification in upper panels and ×1000 magnification in lower panels) from a representative mouse with BCP-ALL. The presence of lymphoblasts in the peripheral blood is obvious, as is the infiltration of the spleen, bone marrow and liver (2nd-4th panels), with effacement of the normal cellular architecture and replacement by nucleolated blasts. B. FACS plots from the bone marrow of a representative mouse demonstrate only background Gr-1/Mac1 myeloid cells, with the majority of cells having a B220+/CD19−/sIg- phenotype in keeping with BCP-ALL. C. D-J PCR rearrangement studies (involving DQ52 and DFL/DSP genes) were performed on spleen or bone marrow gDNA of three mice with phenotypic B-ALL. Negative control was gDNA from C57BL/6J mouse kidney (known to be unrearranged), and positive control was gDNA from a C57BL/6J primary pro-B cell bone marrow culture (known to contain a lot of DJ-rearranged alleles). Sample 1 shows two different rearrangement events (DQ52-J2 and DFL/DSP-J2 rearrangements), in keeping with a clonal B-cell population with both alleles D-J recombined. Sample 2 shows both a germline band and an identical DFL/DSP-J2 band, in keeping with a clonal B-cell population with rearrangement of one allele and germline configuration of the other. Sample 3 shows an absence of recombination.
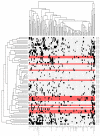
Figure 4. Hierarchical clustering of the leukemias and their mutated genes based on common insertion sites
A matrix of 71 leukemias against the 102 common insertion site (CIS) genes identified by analysis of transposon insertions by the Gaussian kernel convolution method was constructed. Although 73 leukemias were subjected to LM-PCR, sample 120992 did not have any insertions that were mapped and sample 138641 had only 44 insertions sites that were mapped but none of these contributed to any CIS, thus these two samples do not appear in the heatmap as the heatmap is based on the co-occurrence of insertions in CIS. Hierarchical clustering was performed on a binarized version of the matrix using the Hamming distance metric along both the rows and columns, to aggregate leukemias and CIS genes with similar insertions patterns. The two resulting dendrograms were visualized as a heatmap, where black squares indicate the presence of at least one insertion in the tumor, and grey no insertion site. Leukemia rows highlighted in red indicate a BCP-ALL phenotype (n=15; only 14 rows highlighted in red because no insertions were found in the called CIS for sample 138895). The clustering and visualization were performed using the R programming language (version 2.10.0) and Bioconductor (version 2.5).
Figure 5. Insertions in the Ebf1 gene
Three Etv6+/RUNX1; T2Onc+/Tg (EROnc) mice (129753, 132799 and 129713) that developed BCP-ALL carried insertions in the Ebf1 gene (indicated by the red triangles in introns 6 and 8). Sequencing of the insertion-genome junction from splenic cDNA of these mice showed the splicing of Ebf1 directly onto the splice acceptor (SA)-polyA from the transposon. Although sample 129713 carried an insertion in intron 8, we detected splicing onto the transposon directly from exon 6.
Similar articles
- ETV6/RUNX1-like acute lymphoblastic leukemia: A novel B-cell precursor leukemia subtype associated with the CD27/CD44 immunophenotype.
Zaliova M, Kotrova M, Bresolin S, Stuchly J, Stary J, Hrusak O, Te Kronnie G, Trka J, Zuna J, Vaskova M. Zaliova M, et al. Genes Chromosomes Cancer. 2017 Aug;56(8):608-616. doi: 10.1002/gcc.22464. Epub 2017 May 5. Genes Chromosomes Cancer. 2017. PMID: 28395118 - Prevalence of ETV6-RUNX1 fusion gene in children with acute lymphoblastic leukemia in China.
Gao YJ, Zhu XH, Yang Y, Wu Y, Lu FJ, Zhai XW, Wang HS. Gao YJ, et al. Cancer Genet Cytogenet. 2007 Oct 1;178(1):57-60. doi: 10.1016/j.cancergencyto.2007.05.019. Cancer Genet Cytogenet. 2007. PMID: 17889709 - Somatic drivers of B-ALL in a model of ETV6-RUNX1; Pax5(+/-) leukemia.
van der Weyden L, Giotopoulos G, Wong K, Rust AG, Robles-Espinoza CD, Osaki H, Huntly BJ, Adams DJ. van der Weyden L, et al. BMC Cancer. 2015 Aug 13;15:585. doi: 10.1186/s12885-015-1586-1. BMC Cancer. 2015. PMID: 26269126 Free PMC article. - Siblings with ETV6/RUNX1-positive B-lymphoblastic leukemia: A single site experience and review of the literature.
Martig DS, Williamson CM, Xu X, Sukov WR, Greipp PT, Hoppman NL, Baughn LB, Ketterling RP, Peterson JF. Martig DS, et al. Ann Diagn Pathol. 2020 Oct;48:151588. doi: 10.1016/j.anndiagpath.2020.151588. Epub 2020 Aug 14. Ann Diagn Pathol. 2020. PMID: 32836179 Review. - The Landscape of Secondary Genetic Rearrangements in Pediatric Patients with B-Cell Acute Lymphoblastic Leukemia with t(12;21).
Kaczmarska A, Derebas J, Pinkosz M, Niedźwiecki M, Lejman M. Kaczmarska A, et al. Cells. 2023 Jan 18;12(3):357. doi: 10.3390/cells12030357. Cells. 2023. PMID: 36766699 Free PMC article. Review.
Cited by
- Genome-wide interference of ZNF423 with B-lineage transcriptional circuitries in acute lymphoblastic leukemia.
Iglesias P, Puller AC, Seoane M, Spohn M, Raasch S, Klokow M, Müller J, Burkhardt L, Indenbirken D, Horstmann MA. Iglesias P, et al. Blood Adv. 2021 Mar 9;5(5):1209-1223. doi: 10.1182/bloodadvances.2020001844. Blood Adv. 2021. PMID: 33646306 Free PMC article. - Initially disadvantaged, TEL-AML1 cells expand and initiate leukemia in response to irradiation and cooperating mutations.
Li M, Jones L, Gaillard C, Binnewies M, Ochoa R, Garcia E, Lam V, Wei G, Yang W, Lobe C, Hermiston M, Passegué E, Kogan SC. Li M, et al. Leukemia. 2013 Jul;27(7):1570-3. doi: 10.1038/leu.2013.15. Epub 2013 Jan 16. Leukemia. 2013. PMID: 23443342 Free PMC article. No abstract available. - Forward and Reverse Genetics of B Cell Malignancies: From Insertional Mutagenesis to CRISPR-Cas.
Dawes JC, Uren AG. Dawes JC, et al. Front Immunol. 2021 Aug 13;12:670280. doi: 10.3389/fimmu.2021.670280. eCollection 2021. Front Immunol. 2021. PMID: 34484175 Free PMC article. Review. - Sleeping Beauty Mouse Models of Cancer: Microenvironmental Influences on Cancer Genetics.
Guimaraes-Young A, Feddersen CR, Dupuy AJ. Guimaraes-Young A, et al. Front Oncol. 2019 Jul 9;9:611. doi: 10.3389/fonc.2019.00611. eCollection 2019. Front Oncol. 2019. PMID: 31338332 Free PMC article. Review. - Genetic Profiling of Acute and Chronic Leukemia via Next-Generation Sequencing: Current Insights and Future Perspectives.
Pratiwi L, Mashudi FH, Ningtyas MC, Sutanto H, Romadhon PZ. Pratiwi L, et al. Hematol Rep. 2025 Mar 28;17(2):18. doi: 10.3390/hematolrep17020018. Hematol Rep. 2025. PMID: 40277842 Free PMC article. Review.
References
- Romana SP, Mauchauffé M, Le Coniat M, et al. The t(12;21) of acute lymphoblastic leukemia results in a TEL-AML1 gene fusion. Blood. 1995;85:3662–3670. - PubMed
- Shurtleff SA, Buijs A, Behm FG, et al. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. - PubMed
- Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer. 2003;3:639–649. - PubMed
- Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. - PubMed
- Castor A, Nilsson L, Astrand-Grundström I, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11:630–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- A12401/CRUK_/Cancer Research UK/United Kingdom
- 082356/WT_/Wellcome Trust/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 13031/CRUK_/Cancer Research UK/United Kingdom
- G116/187/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases