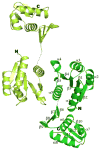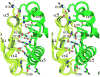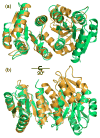Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain - PubMed (original) (raw)
. 2011 Jul 5;50(26):5948-57.
doi: 10.1021/bi2005575. Epub 2011 Jun 13.
Affiliations
- PMID: 21634789
- PMCID: PMC3133661
- DOI: 10.1021/bi2005575
Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain
Smita Menon et al. Biochemistry. 2011.
Erratum in
- Biochemistry. 2011 Aug 16;50(32):7076
Abstract
The PhoP protein from Mycobacterium tuberculosis is a response regulator of the OmpR/PhoB subfamily, whose structure consists of an N-terminal receiver domain and a C-terminal DNA-binding domain. How the DNA-binding activities are regulated by phosphorylation of the receiver domain remains unclear due to a lack of structural information on the full-length proteins. Here we report the crystal structure of the full-length PhoP of M. tuberculosis. Unlike other known structures of full-length proteins of the same subfamily, PhoP forms a dimer through its receiver domain with the dimer interface involving α4-β5-α5, a common interface for activated receiver domain dimers. However, the switch residues, Thr99 and Tyr118, are in a conformation resembling those of nonactivated receiver domains. The Tyr118 side chain is involved in the dimer interface interactions. The receiver domain is tethered to the DNA-binding domain through a flexible linker and does not impose structural constraints on the DNA-binding domain. This structure suggests that phosphorylation likely facilitates/stabilizes receiver domain dimerization, bringing the DNA-binding domains to close proximity, thereby increasing their binding affinity for direct repeat DNA sequences.
Figures
Figure 1
Sequence alignment of response regulators of the OmpR/PhoB subfamily. Sequences were aligned with the program CLUSTALW (49), and manually adjusted based on structural alignments. The secondary structural elements shown are of the PhoP structure. Identical residues are highlighted in black boxes, while residues with high similarity as defined by CLUSTALW are highlighted in gray boxes. The essential aspartate and other acidic residues at the phosphorylation site, conserved lysine (Lys121 in PhoP), and the switch residues (Thr99 and Tyr118) are labeled with asterisks. A vertical arrow marks the boundary between the N- and C-terminal domains. The linker between domains has variable length as indicated by the gaps in the alignment. This figure was generated with the program ALSCRIPT (50).
Figure 2
Ribbon diagram of the full-length PhoP dimer. PhoP dimerizes through α4-β5-α5 of the receiver domain with a two-fold symmetry. The N- and C-termini for both subunits are marked with N and C, respectively. Secondary structural elements for one of the subunits are labeled. The two ends of the disordered linker are connected with a dotted line. This figure and all other color figures were generated with PYMOL (
).
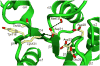
Figure 3
The phosphorylation site and functional residues. The phosphorylation site is located in a shallow pocket at the C-terminal ends of strands β1 and β3. The side chains of acidic residues, a well-conserved Lys121, and switch residues are shown as sticks. Also shown is the carbonyl group of Met73. Thr99 is located at the end of strand β4, and Tyr118 is in the middle of β5. Hydrogen bonds are shown as dashed lines.
Figure 4
A stereo view of the receiver domain dimer interface. Side chains and water molecules not involved in dimer interactions are not shown for clarity. Hydrogen bonds are depicted as dashed lines. A cluster of side chains with flat groups in the center of the interface interact with π-electron stacking, salt bridges, and hydrogen bonds. Most pronounced are side chains of Tyr118 and Arg131, with 2 rings and 2 guanidinium groups stack in parallel with a spacing of ~3.5 Å. There are also two small patches of hydrophobic interactions flanking the central polar interactions.
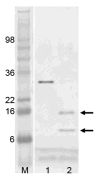
Figure 5
SDS polyacrylamide gel electrophoresis of limited trypsin digestion of PhoP. The lane labeled “M” is the molecular weight marker. Lane 1 is PhoP before trypsin digestion, and lane 2 is after digestion. Calculated molecular weight for PhoP is 27.8 kDa. Protein samples were resolved by SDS-PAGE, and visualized by staining with SimplyBlue. Two bands marked with arrows are sequenced by Edman degradation to identify trypsin cleavage sites.
Figure 6
Structural alignment of the receiver domain dimer of PhoP with that of activated PhoB from E. coli (PDB code 1ZES). The PhoP structure is colored in green, and PhoB is colored in orange. The structural superposition was carried out on one of the subunits, shown on the left, with 111 matched Cα atoms and an rmsd of 1.5 Å. The other subunit, shown on the right, has a shift between the two proteins. Panel (a) has the two-fold symmetric axis of the PhoP dimer perpendicular to the paper similar to the view in Figure 2, and panel (b) is a 90° rotation from the view in (a). Despite that both involve α4-β5-α5, the interactions at the dimer interface are different in PhoP and PhoB. The activated PhoB dimer has a more compact interface, with helices from subunits packing more closely.
Similar articles
- Atypical OmpR/PhoB subfamily response regulator GlnR of actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions.
Lin W, Wang Y, Han X, Zhang Z, Wang C, Wang J, Yang H, Lu Y, Jiang W, Zhao GP, Zhang P. Lin W, et al. J Biol Chem. 2014 May 30;289(22):15413-25. doi: 10.1074/jbc.M113.543504. Epub 2014 Apr 14. J Biol Chem. 2014. PMID: 24733389 Free PMC article. - Structural basis of DNA sequence recognition by the response regulator PhoP in Mycobacterium tuberculosis.
He X, Wang L, Wang S. He X, et al. Sci Rep. 2016 Apr 15;6:24442. doi: 10.1038/srep24442. Sci Rep. 2016. PMID: 27079268 Free PMC article. - The crystal structure of the phosphorylation domain in PhoP reveals a functional tandem association mediated by an asymmetric interface.
Birck C, Chen Y, Hulett FM, Samama JP. Birck C, et al. J Bacteriol. 2003 Jan;185(1):254-61. doi: 10.1128/JB.185.1.254-261.2003. J Bacteriol. 2003. PMID: 12486062 Free PMC article. - The structure of a full-length response regulator from Mycobacterium tuberculosis in a stabilized three-dimensional domain-swapped, activated state.
King-Scott J, Nowak E, Mylonas E, Panjikar S, Roessle M, Svergun DI, Tucker PA. King-Scott J, et al. J Biol Chem. 2007 Dec 28;282(52):37717-29. doi: 10.1074/jbc.M705081200. Epub 2007 Oct 16. J Biol Chem. 2007. PMID: 17942407 - Molecular strategies for phosphorylation-mediated regulation of response regulator activity.
Gao R, Stock AM. Gao R, et al. Curr Opin Microbiol. 2010 Apr;13(2):160-7. doi: 10.1016/j.mib.2009.12.009. Epub 2010 Jan 14. Curr Opin Microbiol. 2010. PMID: 20080056 Free PMC article. Review.
Cited by
- Bordetella pertussis fim3 gene regulation by BvgA: phosphorylation controls the formation of inactive vs. active transcription complexes.
Boulanger A, Moon K, Decker KB, Chen Q, Knipling L, Stibitz S, Hinton DM. Boulanger A, et al. Proc Natl Acad Sci U S A. 2015 Feb 10;112(6):E526-35. doi: 10.1073/pnas.1421045112. Epub 2015 Jan 26. Proc Natl Acad Sci U S A. 2015. PMID: 25624471 Free PMC article. - Identification of drivers of mycobacterial resistance to peptidoglycan synthesis inhibitors.
Olivença F, Ferreira C, Nunes A, Silveiro C, Pimentel M, Gomes JP, Catalão MJ. Olivença F, et al. Front Microbiol. 2022 Sep 6;13:985871. doi: 10.3389/fmicb.2022.985871. eCollection 2022. Front Microbiol. 2022. PMID: 36147841 Free PMC article. - Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Khosa S, Hoeppner A, Gohlke H, Schmitt L, Smits SH. Khosa S, et al. PLoS One. 2016 Mar 1;11(3):e0149903. doi: 10.1371/journal.pone.0149903. eCollection 2016. PLoS One. 2016. PMID: 26930060 Free PMC article. - Solution structure of the PhoP DNA-binding domain from Mycobacterium tuberculosis.
Macdonald R, Sarkar D, Amer BR, Clubb RT. Macdonald R, et al. J Biomol NMR. 2015 Sep;63(1):111-7. doi: 10.1007/s10858-015-9965-0. Epub 2015 Jul 25. J Biomol NMR. 2015. PMID: 26209027 Free PMC article. - Structure of the Acinetobacter baumannii PmrA receiver domain and insights into clinical mutants affecting DNA binding and promoting colistin resistance.
Palethorpe S, Milton ME, Pesci EC, Cavanagh J. Palethorpe S, et al. J Biochem. 2022 Jan 7;170(6):787-800. doi: 10.1093/jb/mvab102. J Biochem. 2022. PMID: 34585233 Free PMC article.
References
- Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr Opin Microbiol. 2010;13:232–239. - PubMed
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
- Tucker PA, Nowak E, Morth JP. Two-component systems of Mycobacterium tuberculosis: structure-based approaches. Methods Enzymol. 2007;423:479–501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources