The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication - PubMed (original) (raw)
The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication
Kimberly L Carey et al. PLoS Pathog. 2011 May.
Abstract
Coxiella burnetii, the causative agent of human Q fever, is an intracellular pathogen that replicates in an acidified vacuole derived from the host lysosomal network. This pathogen encodes a Dot/Icm type IV secretion system that delivers bacterial proteins called effectors to the host cytosol. To identify new effector proteins, the functionally analogous Legionella pneumophila Dot/Icm system was used in a genetic screen to identify fragments of C. burnetii genomic DNA that when fused to an adenylate cyclase reporter were capable of directing Dot/Icm-dependent translocation of the fusion protein into mammalian host cells. This screen identified Dot/Icm effectors that were proteins unique to C. burnetii, having no overall sequence homology with L. pneumophila Dot/Icm effectors. A comparison of C. burnetii genome sequences from different isolates revealed diversity in the size and distribution of the genes encoding many of these effectors. Studies examining the localization and function of effectors in eukaryotic cells provided evidence that several of these proteins have an affinity for specific host organelles and can disrupt cellular functions. The identification of a transposon insertion mutation that disrupts the dot/icm locus was used to validate that this apparatus was essential for translocation of effectors. Importantly, this C. burnetii Dot/Icm-deficient mutant was found to be defective for intracellular replication. Thus, these data indicate that C. burnetii encodes a unique subset of bacterial effector proteins translocated into host cells by the Dot/Icm apparatus, and that the cumulative activities exerted by these effectors enables C. burnetii to successfully establish a niche inside mammalian cells that supports intracellular replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
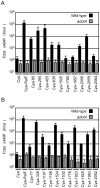
Figure 1. Dot/Icm-dependent translocation of C. burnetii proteins by L. pneumophila.
CHO-FcγRII cells were infected with Dot/Icm-sufficient LP01 strain of L. pneumophila (black) or the isogenic Δ_dotA_ mutant (grey) expressing Cya fusions to the indicated C. burnetii proteins. (A) Fusions to Cya of the indicated full-length derivatives of C. burnetii NM proteins identified in the genetic screen were tested for translocation (B). Fusions to Cya of the indicated full-length derivatives of C. burnetii NM proteins identified based on homology or proximity to proteins identified in the screen were tested for translocation. Cya indicates empty vector control. Cya-RalF was used as a positive control. After 1 h, host cells were lysed and cAMP was extracted. Total cAMP levels resulting from translocation of protein fusions were quantified using an enzyme-immunoassay system, and are shown in fmol. Results represent average values +/− SD of experiments performed in triplicate.
Figure 2. Domain analysis and genomic comparisons for C. burnetii effectors identified in this study.
The schematic shows the eighteen NM effectors identified in this study represented in blue. Below each NM effector is a representation of the size of the homologous reading frame encoded by the genes in the other sequenced strains of C. burnetii. The size of the predicted proteins is represented in kDa in brackets following the gene designation. Black lines represent the presence of homologous DNA that does not encode an open reading frame due to small deletions (vertical black line) and stop codons (red cross). In cases where multiple deletions occur the first deletion, representing the site of the frameshift, is displayed. The locations of conserved domains in each protein identified in a SMART database search are indicated according to the key provided.
Figure 3. Effectors of C. burnetii show differential localization patterns when expressed in mammalian HeLa 229 cells.
Immunofluorescence micrographs show the representative localization profiles of the indicated FLAG-tagged C. burnetii effectors (Green) and the indicated host organelles (Red) after ectopic production of the proteins in HeLa 229 cells. (A) CbuK1976 and CBU1524 show localization to the nucleus, but the NM protein CBU0080 is distributed throughout the cytoplasm. The schematic shows that CBU0080 does not contain the helix-loop-helix motif (octagon) found in the homolog CbuK1976, which could explain the differential localization phenotypes observed (B) Cells stained for FLAG-tagged CBU0077 and LAMP1 show colocalization of the effector on lysosome-derived organelles, and cells stained for CBU0635 and GM130 show distribution of the protein to a perinuclear region containing the Golgi apparatus. (C). Cells stained for FLAG-tagged effectors and the TOM22 protein show colocalization of the effectors on mitochondria. The NM protein CBU1825 and the homologous Dugway protein CBUD0054 showed similar localization patterns. The NM protein CBU1532 and the Dugway homolog CBUD0454 showed slightly different mitochondrial straining patterns, with CBU1532 production leading to cell rounding and mitochondria aggregation. Scale bars represent 10 µm.
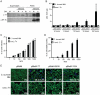
Figure 4. CBU0635 interferes with host protein secretion.
The impact of C. burnetii Dot/Icm effectors on the host cell secretory pathway was examined by monitoring secretion of the SEAP protein into the culture supernatant. (A) HEK 293 cells were co-transfected with pSEAP and the plasmid encoding the indicated effector protein or the GDP-locked ARF1T31N protein (pArf1-T31N) or empty vector (pFLAG). External and internal SEAP activity was measured. The grey bars show that there was a significant decrease in the external/internal ratio of SEAP activity observed upon ectopic expression of ARF1T31N (P = 0.0012) or CBU0635 (P = 0.0057) in comparison to HEK 293 cells transfected with pFLAG alone (black bar). Expression of all other C. burnetii effectors did not significantly the ratio of SEAP activity (white bars). (B). SEAP activity (y-axis) was measured at the indicated times after cells were washed and new culture medium was added (x-axis) Data show SEAP ratios for cells with vector alone (pFLAG; black triangles), cells producing CBU0635 (pCBU0635; open squares) and cells producing ARF1T31N (grey circles). A similar defect in SEAP secretion was observed in cells producing ARF1T31N as in cells producing CBU0635.
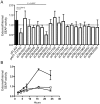
Figure 5. Several C. burnetii effectors slow yeast replication.
(A) Yeast strains were grown in YNB supplemented with 2% galactose to induce the expression of the indicated effector proteins listed in the legend on the right. The plots show yeast replication as determined by measuring the optical density of the culture at 600 nm (OD 600 nm, y-axis) every 30 min (x-axis) for a period of 36 h. Growth of S. cerevisiae expressing C. burnetii effectors were compared to controls that included S. cerevisiae containing the vector control (pYES2; open squares) and S. cerevisiae producing the L. pneumophila effector YlfA (pJG4-5:YlfA; red triangles). Effectors that resulted in a delay in the doubling rate of S. cerevisiae are highlighted with red symbols. (B) The doubling time of S. cerevisiae producing the indicated effector proteins was calculated during the exponential phase of growth from the growth curves shown in panel A. Effectors that resulted in a significant increase in doubling time compared to S. cerevisiae pYES2 are highlighted in open boxes (P<0.01). Data represent the mean doubling time ± SD determined from at least 3 independent growth curves.
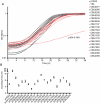
Figure 6. The C. burnetii Dot/Icm system is essential for intracellular replication.
(A) A clone of C. burnetii with a 3.69 kb transposon inserted in the icmL gene (dotI/icmL.1) was isolated after enrichment on medium containing kanamycin. The schematic shows the predicted location and orientation of the transposon in icmL as determined by sequence analysis. The location of probes and restriction sites used to validate the site of transposon insertion by PCR and Southern hybridization are indicated. (B) As indicated, genomic DNA digested with _Hind_III isolated from the C. burnetii NM strain and the isogenic icmL::Tn mutant was analyzed by Southern hybridization using a probe derived from the icmL gene and a probe derived from the transposon. As predicted from the schematic in panel A, the icmL probe identified a single band of 4.2 kb in the NM strain, and two bands of 3.3 kb and 2.5 kb in the icmL::Tn mutant because of a new _Hind_III site introduced into the icmL locus by the transposon. When a probe homologous to the transposon was used (Tn probe), a single band was identified in the icmL::Tn mutant, indicating that this strain has a single insertion of the transposon in the chromosome. (C) PCR amplification of genomic DNA from NM and the icmL::Tn mutant using primer sets shown in panel A confirm the predicted location and orientation of the transposon insertion in icmL, and that the transposon is retained by the icmL::Tn strain after introduction of a plasmid encoding icmL (pIcmL) or a plasmid encoding the entire operon harboring icmL (pQC). (D,E) The ability of the icmL::Tn mutant to replicate in HeLa (D) and Vero (E) cells was determined by measuring genomic equivalents (y-axis) at the indicated times after infection (x-axis). There was a 50 to 100-fold increase over 7 days in C. burnetii NM genomic units (black squares). No significant increase in genomic units was detected for the icmL::Tn mutant (white squares) or C. burnetii NM treated with 10 µg/ml chloramphenicol (gray squares). (F,G) The ability of the icmL::Tn mutant to replicate in HeLa (F) and Vero (G) cells was determined by measuring genomic equivalents (y-axis) at the indicated times after infection (x-axis). Replication was observed for C. burnetii NM containing pBlaM (black squares) and the icmL::Tn mutant containing the plasmid pQC (black triangles). The icmL::Tn mutant containing the vector pBlaM (white squares) or for the icmL::Tn mutant containing pIcmL (white triangles). Graphs represent the mean ± SD of at least three independent experiments. (H) Fluorescence micrographs were acquired after infection of HeLa cells for 5 days by the strains of C. burnetii indicated. An anti-Coxiella antibody (green) was used to visualize intracellular bacteria and the nucleus of the host cell was visualized by DAPI staining (blue). Replicating bacteria in large vacuoles were observed for cells infected with C. burnetii NM, C. burnetii NM containing pBlaM and the icmL::Tn mutant containing pQC. Only individual bacteria were observed inside the host cells infected with the icmL::Tn mutant, C. burnetii NM in medium with chloramphenicol, icmL::Tn containing pBlaM and icmL::Tn containing pIcmL (indicated with arrows). These are representative images from at least three independent experiments.
Figure 7. The C. burnetii Dot/Icm secretion system is required for effector translocation.
(A) HeLa cells either infected with C. burnetii NM or uninfected were incubated with or without chloramphenicol (CM) and cellular lysates were separated into supernatant and pellet fractions. Immunoblot analysis was used to detect CBU0077 (top panel; α-77) and EF-Ts (bottom panel; α-EF-Ts). The CBU0077 protein was detected in both the supernatant fraction containing proteins secreted during infection and in the pellet fraction containing intact bacteria. The cell associated EF-Ts protein was detected only in the pellet fraction containing intact bacterial cells. The addition of chloramphenicol resulted in a slight drop in levels of CBU0077 detected in the supernatant, suggesting the secreted protein has a relatively long half-life. Similar results were obtained from three independent experiments. (B) C. burnetii NM (black bars) and the icmL::Tn mutant (white bars) containing either pBlaM or pBlaM-77 were used to infect HeLa cells for the times indicated (x-axis). Translocation of the BlaM and BlaM-CBU0077 fusion protein by the respective strains was determined by measuring the change in the 460 nm/535 nm fluorescence emission ratio resulting from cleavage of the CCF4-AM substrate (y-axis). Results represent the mean ± SD obtained from triplicate samples. (C, D) HeLa cells were infected in parallel with C. burnetii NM producing BlaM-CBU0077 or the icmL::Tn strain producing BlaM-CBU0077 at the multiplicities of infection (MOI) indicated on the x-axis. Fluorescence microscopy was used to determine the percent of cells infected by the two strains (C) and the number of cells that stained positive for BlaM translocation (D) Results represent the mean ± SD obtained from triplicate samples. (E) Fluorescence micrographs of HeLa cells that were infected for 24 h with either C. burnetii NM or the icmL::Tn strains producing either BlaM alone or BlaM fusions to CBU0077 (pBlaM-77), CBU0635 (pBlaM-0635) or CBU1524 (pBlaM-1524). Emission at 535 nm (green) identifies the CCF4-AM-loaded cells and emission at 460 nm (blue) indicates cleavage of CCF4-AM in the cytosol of the infected cell resulting from translocation of the BlaM fusion protein. The percent of cells that were BlaM positive (blue) was determined for each infection condition and the mean ± SD from three independent experiments is displayed in the bottom right corner of each representative micrograph.
Similar articles
- Dependency of Coxiella burnetii Type 4B Secretion on the Chaperone IcmS.
Larson CL, Beare PA, Heinzen RA. Larson CL, et al. J Bacteriol. 2019 Nov 5;201(23):e00431-19. doi: 10.1128/JB.00431-19. Print 2019 Dec 1. J Bacteriol. 2019. PMID: 31501284 Free PMC article. - Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole.
Newton HJ, McDonough JA, Roy CR. Newton HJ, et al. PLoS One. 2013;8(1):e54566. doi: 10.1371/journal.pone.0054566. Epub 2013 Jan 17. PLoS One. 2013. PMID: 23349930 Free PMC article. - Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system.
Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Zamboni DS, et al. Mol Microbiol. 2003 Aug;49(4):965-76. doi: 10.1046/j.1365-2958.2003.03626.x. Mol Microbiol. 2003. PMID: 12890021 - Coxiella burnetii secretion systems.
McDonough JA, Newton HJ, Roy CR. McDonough JA, et al. Adv Exp Med Biol. 2012;984:171-97. doi: 10.1007/978-94-007-4315-1_9. Adv Exp Med Biol. 2012. PMID: 22711632 Review. - Legionella and Coxiella effectors: strength in diversity and activity.
Qiu J, Luo ZQ. Qiu J, et al. Nat Rev Microbiol. 2017 Oct;15(10):591-605. doi: 10.1038/nrmicro.2017.67. Epub 2017 Jul 17. Nat Rev Microbiol. 2017. PMID: 28713154 Review.
Cited by
- Autophagy and bacterial infection: an evolving arms race.
Choy A, Roy CR. Choy A, et al. Trends Microbiol. 2013 Sep;21(9):451-6. doi: 10.1016/j.tim.2013.06.009. Epub 2013 Jul 20. Trends Microbiol. 2013. PMID: 23880062 Free PMC article. - Murine Alveolar Macrophages Are Highly Susceptible to Replication of Coxiella burnetii Phase II In Vitro.
Fernandes TD, Cunha LD, Ribeiro JM, Massis LM, Lima-Junior DS, Newton HJ, Zamboni DS. Fernandes TD, et al. Infect Immun. 2016 Aug 19;84(9):2439-48. doi: 10.1128/IAI.00411-16. Print 2016 Sep. Infect Immun. 2016. PMID: 27297388 Free PMC article. - Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages.
Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. Graham JG, et al. Cell Microbiol. 2013 Jun;15(6):1012-25. doi: 10.1111/cmi.12096. Epub 2013 Jan 14. Cell Microbiol. 2013. PMID: 23279051 Free PMC article. - The Coxiella burnetii T4SS effector protein AnkG hijacks the 7SK small nuclear ribonucleoprotein complex for reprogramming host cell transcription.
Cordsmeier A, Rinkel S, Jeninga M, Schulze-Luehrmann J, Ölke M, Schmid B, Hasler D, Meister G, Häcker G, Petter M, Beare PA, Lührmann A. Cordsmeier A, et al. PLoS Pathog. 2022 Feb 8;18(2):e1010266. doi: 10.1371/journal.ppat.1010266. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35134097 Free PMC article. - Syntaxin 11 Contributes to the Interferon-Inducible Restriction of Coxiella burnetii Intracellular Infection.
Ganesan S, Alvarez NN, Steiner S, Fowler KM, Corona AK, Roy CR. Ganesan S, et al. mBio. 2023 Feb 28;14(1):e0354522. doi: 10.1128/mbio.03545-22. Epub 2023 Feb 2. mBio. 2023. PMID: 36728431 Free PMC article.
References
- Howe D, Heinzen RA. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol. 2006;8:496–507. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- F32 AI066547/AI/NIAID NIH HHS/United States
- R01 AI041699-15/AI/NIAID NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
- R01 AI064559-05/AI/NIAID NIH HHS/United States
- R01 AI041699/AI/NIAID NIH HHS/United States
- AI066547/AI/NIAID NIH HHS/United States
- R01 AI064559/AI/NIAID NIH HHS/United States
- R01-AI064559/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources