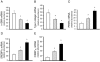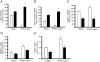Adiponectin regulation of stellate cell activation via PPARγ-dependent and -independent mechanisms - PubMed (original) (raw)
Adiponectin regulation of stellate cell activation via PPARγ-dependent and -independent mechanisms
Mahnoush S Shafiei et al. Am J Pathol. 2011 Jun.
Abstract
In this study, we elucidated the mechanism by which adiponectin modulates hepatic stellate cell activation and fibrogenesis. Adiponectin-overexpressing transgenic mice receiving thioacetamide were resistant to fibrosis, compared with controls. In contrast, adiponectin-null animals developed severe fibrosis. Expression of collagen α1(I) and α-smooth muscle actin (α-SMA) mRNAs were significantly lower in adiponectin-overexpressing mice, compared with controls. In wild-type stellate cells exposed to a lentivirus encoding adiponectin, expression of peroxisome proliferator-activated receptor-γ (PPARγ), SREBP1c, and CEBPα mRNAs was significantly increased (3.2-, 4.1-, and 2.2-fold, respectively; n = 3; P < 0.05, adiponectin virus versus control), consistent with possible activation of an adipogenic transcriptional program. Troglitazone, a PPARγ agonist, strongly suppressed up-regulation of collagen α1(I) and α-SMA mRNA in stellate cells isolated from wild-type mice; however, stellate cells from adiponectin-null animals failed to respond to troglitazone. Furthermore, in isolated stellate cells in which PPARγ was depleted using an adenovirus-Cre-recombinase system and in which adiponectin was also overexpressed, collagen α1(I) and α-SMA were significantly inhibited. We conclude that the PPARγ effect on stellate cell activation and the fibrogenic cascade appears to be adiponectin-dependent; however, the inhibitory effect of adiponectin on stellate cell activation was not dependent on PPARγ, suggesting the presence of PPARγ-dependent as well as independent pathways in stellate cells.
Copyright © 2011 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures
Figure 1
Characterization of stellate cells derived from adiponectin-deficient and transgenic mice. A: Stellate cells were isolated from wild type (WT), adiponectin-knockout (KO), and adiponectin-transgenic (Tg) mice and were grown in 20% serum-containing medium for up to 2 days. Representative images are shown (n > 10). Scale bar = 10 μm. B and D: mRNA levels of α-SMA (B) and collagen α1(I) (D) were measured after 7 days in culture (n = 4). *P < 0.05 versus WT; **P < 0.05 versus KO. C: Immunoblotting was performed to detect α-SMA and data were quantitated (means ± SE; n = 4). *P < 0.05 versus WT.
Figure 2
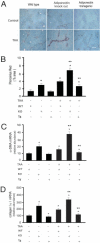
Liver morphology and inflammation in adiponectin-deficient and transgenic mice. Liver fibrosis was induced with repetitive intraperitoneal injection of 0.2 g/kg body weight of thioacetamide. At the end of the experimental period, liver samples were collected for histological and biochemical assays. A: Representative images of liver sections stained with H&E. Areas in insets are shown in higher magnification in the bottom row. Scale bars = 50 μm. B: mRNA was isolated from liver tissue, and relative abundance of F4/80 mRNA levels was measured by RT-PCR (means ± SE; n = 4). *P < 0.05 versus WT; **P < 0.05 versus KO.
Figure 3
Mice lacking adiponectin are susceptible to fibrosis and mice overexpressing adiponectin are resistant to fibrosis. Liver fibrosis was induced by repetitive intraperitoneal injection of 0.2 g/kg body weight of thioacetamide (TAA). At the end of the experimental period, liver samples were collected for histological and biochemical examinations. A: Liver sections were stained with picrosirius red. Scale bar = 50 μm. B: Histomorphometric analysis was performed on random picrosirius red-stained liver sections (means ± SE; n = 10 fields/liver and 10 livers/group). *P < 0.05 versus WT alone; **P < 0.05 versus TAA alone. C and D: Livers were harvested, total RNA was extracted, and real-time PCR was performed to detect mRNA expression of α-SMA (C) and collagen α1(I) (D) (means ± SE; n = 6). *P < 0.05 versus WT control; **P < 0.05 versus WT and TAA.
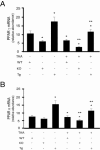
Figure 4
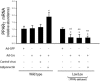
PPAR expression in adiponectin-deficient and transgenic mice. Liver fibrosis was induced by repetitive intraperitoneal injection of 0.2 g/kg body weight of thioacetamide (TAA). Livers were harvested, total RNA was extracted, and real-time PCR was performed to detect mRNA expression of PPARγ (A) or PPARα (B) (means ± SE; n = 6). *P < 0.05 versus WT; **P < 0.05 versus WT and TAA.
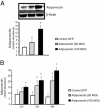
Figure 5
Quantification of adiponectin protein expression in hepatic stellate cells. Stellate cells from WT mice on FVB background were isolated, cultured for up to 48 hours, and then infected with a lentivirus expressing GFP, or adiponectin at 50 and 100 MOI. Cell lysates were harvested at 48 hours (A) or at 12, 24, and 48 hours (B) and were subjected to immunoblotting. A representative immunoblot is shown (A). Subsequently, specific bands from repeated experiments were quantified and normalized to the signal for β-actin (A and B) (means ± SE; n = 3). *P < 0.05 versus control GFP.
Figure 6
Effects of adiponectin overexpression on stellate cell activation. Stellate cells from normal rats were isolated, plated at equivalent density, and allowed to undergo culture-induced activation for 3 days. Cells were transduced with a control lentivirus expressing GFP or lentivirus overexpressing adiponectin at 50 and 100 MOI (n = 3/group). Total RNA was harvested 72 hours later and was subjected to RT-PCR to detect mRNA expression of α-SMA (A), collagen α1(I) (B), PPARγ (C), SREBP1c (D), and CEBPα (E) (means ± SE; n = 3). White bars indicate control GFP; gray bars indicate adiponectin (50 MOI); black bars indicate adiponectin (100 MOI). *P < 0.05 versus control GFP.
Figure 7
Adiponectin is required for PPARγ ligands to be effective in inhibiting stellate cell activation. Stellate cells isolated from WT and adiponectin knockout (ADNKO) mice were cultured in the presence of serum-containing medium for 24 hours. Cells were exposed to 5 μg/mL of the PPARγ ligand troglitazone, or an equivalent volume of dimethyl sulfoxide (vehicle) for 4 days, after which total RNA was extracted from lysates and mRNA expression of α-SMA (A), collagen α1(I) (B), CEBPα (C), SREBP1c (D), and PPARγ (E) was quantified by real-time PCR (means ± SE; n = 3). White bars indicate WT; black bars indicate ADNKO. *P < 0.05 versus WT cells exposed to vehicle alone; **P < 0.05 versus WT cells exposed to troglitazone.
Figure 8
Deletion of PPARγ with a Cre-expressing adenovirus. Stellate cells were isolated from WT and PPARγ Cre/Lox mice and grown in culture. After 48 hours, cells were infected with adenovirus expressing Cre and/or lentivirus encoding adiponectin or corresponding control for 4 days. Total cellular RNA was isolated and mRNA expression of PPARγ was quantified by RT-PCR (means ± SE; n = 3). *P < 0.05 versus WT cells exposed to adenovirus expressing GFP alone; **P < 0.05 versus Lox/Lox cells exposed to adenovirus expressing GFP alone.
Figure 9
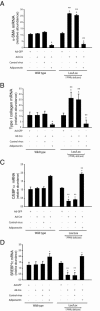
The antiactivating and antifibrotic actions of adiponectin are independent of PPARγ. Stellate cells were isolated from WT and PPARγ Cre/Lox mice and grown in culture. After 48 hours, cells were infected with Cre-expressing adenovirus and/or lentivirus encoding adiponectin or corresponding control for four days. Total cellular RNA was isolated and mRNA expression of α-SMA (A), collagen α1(I) (B), CEBPα (C), and SREBP1c (D) was quantified by RT-PCR (means ± SE; n = 3). *P < 0.05 versus WT cells exposed to adenovirus expressing GFP alone; **P < 0.05 versus Lox/Lox cells exposed to adenovirus expressing GFP alone.
Similar articles
- Regulation of peroxisome proliferator-activated receptor-gamma in liver fibrosis.
Yang L, Chan CC, Kwon OS, Liu S, McGhee J, Stimpson SA, Chen LZ, Harrington WW, Symonds WT, Rockey DC. Yang L, et al. Am J Physiol Gastrointest Liver Physiol. 2006 Nov;291(5):G902-11. doi: 10.1152/ajpgi.00124.2006. Epub 2006 Jun 22. Am J Physiol Gastrointest Liver Physiol. 2006. PMID: 16798724 - Effectiveness of the PPARγ agonist, GW570, in liver fibrosis.
Yang L, Stimpson SA, Chen L, Wallace Harrington W, Rockey DC. Yang L, et al. Inflamm Res. 2010 Dec;59(12):1061-71. doi: 10.1007/s00011-010-0226-0. Epub 2010 Jun 29. Inflamm Res. 2010. PMID: 20585829 - Histone H3K9 demethylase JMJD1A modulates hepatic stellate cells activation and liver fibrosis by epigenetically regulating peroxisome proliferator-activated receptor γ.
Jiang Y, Wang S, Zhao Y, Lin C, Zhong F, Jin L, He F, Wang H. Jiang Y, et al. FASEB J. 2015 May;29(5):1830-41. doi: 10.1096/fj.14-251751. Epub 2015 Jan 21. FASEB J. 2015. PMID: 25609425 - Peroxisome proliferator-activated receptor-γ as a therapeutic target for hepatic fibrosis: from bench to bedside.
Zhang F, Kong D, Lu Y, Zheng S. Zhang F, et al. Cell Mol Life Sci. 2013 Jan;70(2):259-76. doi: 10.1007/s00018-012-1046-x. Epub 2012 Jun 15. Cell Mol Life Sci. 2013. PMID: 22699820 Free PMC article. Review. - Peroxisome proliferator-activated receptor-γ cross-regulation of signaling events implicated in liver fibrogenesis.
Zhang F, Lu Y, Zheng S. Zhang F, et al. Cell Signal. 2012 Mar;24(3):596-605. doi: 10.1016/j.cellsig.2011.11.008. Epub 2011 Nov 13. Cell Signal. 2012. PMID: 22108088 Review.
Cited by
- Pegbelfermin for reducing transaminase levels in patients with non-alcoholic steatohepatitis: a dose-response meta-analysis of randomized controlled trials.
Lu Y, Yu B, Bu Y, Lou J, Jin Y. Lu Y, et al. Front Med (Lausanne). 2024 Apr 5;11:1293336. doi: 10.3389/fmed.2024.1293336. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38646552 Free PMC article. - Adiponectin: friend or foe in obesity and inflammation.
Luo L, Liu M. Luo L, et al. Med Rev (2021). 2022 Jul 1;2(4):349-362. doi: 10.1515/mr-2022-0002. eCollection 2022 Aug. Med Rev (2021). 2022. PMID: 37724325 Free PMC article. Review. - Stromal inflammation, fibrosis and cancer: An old intuition with promising potential.
Oey O, Sunjaya AF, Khan Y, Redfern A. Oey O, et al. World J Clin Oncol. 2023 Jul 24;14(7):230-246. doi: 10.5306/wjco.v14.i7.230. World J Clin Oncol. 2023. PMID: 37583950 Free PMC article. Review. - Perilipin 2 Protects against Lipotoxicity-Induced Islet Fibrosis by Inducing Islet Stellate Cell Activation Phenotype Changes.
Zhou Y, Wang Y, Ni C, Wang H, Zhou J, Wan B, Li H, Li F, Huang R, Xu W, Shan T, Cai T, Kong X, Liu B, Liu X, Sun Z, Ma J. Zhou Y, et al. Biomed Res Int. 2022 Jul 6;2022:4581405. doi: 10.1155/2022/4581405. eCollection 2022. Biomed Res Int. 2022. PMID: 35845956 Free PMC article. - Adiponectin preserves metabolic fitness during aging.
Li N, Zhao S, Zhang Z, Zhu Y, Gliniak CM, Vishvanath L, An YA, Wang MY, Deng Y, Zhu Q, Shan B, Sherwood A, Onodera T, Oz OK, Gordillo R, Gupta RK, Liu M, Horvath TL, Dixit VD, Scherer PE. Li N, et al. Elife. 2021 Apr 27;10:e65108. doi: 10.7554/eLife.65108. Elife. 2021. PMID: 33904399 Free PMC article.
References
- Kamada Y., Takehara T., Hayashi N. Adipocytokines and liver disease. J Gastroenterol. 2008;43:811–822. - PubMed
- Rombouts K., Marra F. Molecular mechanisms of hepatic fibrosis in non-alcoholic steatohepatitis. Dig Dis. 2010;28:229–235. - PubMed
- Farrell G.C., Larter C.Z. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. - PubMed
- Friedman S.L., Rockey D.C., Bissell D.M. Hepatic fibrosis 2006: report of the Third AASLD Single Topic Conference. Hepatology. 2007;45:242–249. - PubMed
- Tsukamoto H., She H., Hazra S., Cheng J., Miyahara T. Anti-adipogenic regulation underlies hepatic stellate cell transdifferentiation. J Gastroenterol Hepatol. 2006;21(Suppl 3):S102–S105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 DK088761/DK/NIDDK NIH HHS/United States
- RC1 DK086629/DK/NIDDK NIH HHS/United States
- R01-DK50574/DK/NIDDK NIH HHS/United States
- R01-DK55758/DK/NIDDK NIH HHS/United States
- R01-CA112023/CA/NCI NIH HHS/United States
- R01 DK050574/DK/NIDDK NIH HHS/United States
- R01 DK055758/DK/NIDDK NIH HHS/United States
- R01 CA112023/CA/NCI NIH HHS/United States
- RC1-DK086629/DK/NIDDK NIH HHS/United States
- P01-DK088761/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases