Improved technologies now routinely provide protein NMR structures useful for molecular replacement - PubMed (original) (raw)
Improved technologies now routinely provide protein NMR structures useful for molecular replacement
Binchen Mao et al. Structure. 2011.
Abstract
Molecular replacement (MR) is widely used for addressing the phase problem in X-ray crystallography. Historically, crystallographers have had limited success using NMR structures as MR search models. Here, we report a comprehensive investigation of the utility of protein NMR ensembles as MR search models, using data for 25 pairs of X-ray and NMR structures solved and refined using modern NMR methods. Starting from NMR ensembles prepared by an improved protocol, FindCore, correct MR solutions were obtained for 22 targets. Based on these solutions, automatic model rebuilding could be done successfully. Rosetta refinement of NMR structures provided MR solutions for another two proteins. We also demonstrate that such properly prepared NMR ensembles and X-ray crystal structures have similar performance when used as MR search models for homologous structures, particularly for targets with sequence identity >40%.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
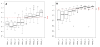
Fig. 1. Knowledge-based structure quality scores for NESG NMR structures have consistently improved as NMR methods have matured over the past several years
Panel A and B show boxplots of the distribution of Z scores (y-axis) of Procheck ‘all-dihedral-angle’ G-factor and Molprobity clashscores, respectively, for all NMR structures solved by the NESG consortium in each PSI fiscal year (x-axis). The red dashed lines represent the average Z scores. One PSI fiscal year is a 12 month time period generally spanning July 1st through June 30th of the following year. The Procheck all-dihedral-angle G-factor is determined by the stereochemical quality of both backbone and side chain dihedral angles of proteins, and Molprobity clashscore is a measure to reflect the number of high-energy contacts in a structure calculated by the program probe. PSVS Z scores are calculated based on a calibrated dataset of 252 high quality X-ray crystal structures from the PDB with resolution ≤ 1.80 Å, R-factor ≤ 0.25, and R-free ≤ 0.28 (Bhattacharya et al. 2007).
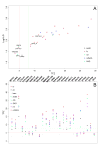
Fig. 2. Using the fc method, Phaser phasing scores obtained using NMR structure ensembles as templates are generally sufficient to provide good MR solutions
(A) LLG-TFZ scatter plot. LLG (log-likelihood gain) and TFZ (translation function Z-score) scores are calculated by Phaser, and log10(LLG) and TFZ scores are plotted on y-axis and x-axis respectively. The red vertical dash line delimits (TFZ=5), the typical cut-off of an invalid Phaser solution, while the green vertical dash line (TFZ=8) delimits the typical cut-off of a definite Phaser solution according to the Phaser manual. For each individual target, only the model with the highest TFZ score solution is plotted. Colors are coded by different model preparation methods. SR478_R and ZR18_R denote the two models following Rosetta refinement. (B) Comparisons of TFZ scores from different MR models prepared by the eight model preparation methods. Models are color coded by their respective preparation method. TFZ scores calculated by Phaser are plotted on y-axis, while each NESG target is plotted on x-axis in alphabetical order. The red horizontal dash line (at TFZ=5) delimits the typical cut-off of an invalid Phaser solution while the green horizontal dash line (at TFZ=8) delimits the typical cut-off of a definite Phaser solution, according to the Phaser Manual. See also Tables S2 and S5.
Fig. 3. NMR and X-ray structures are about equally useful as templates for obtaining MR solutions for homologous protein structures
(A) Plot of TFZ scores of Phaser solutions vs. sequence identity (Seq_ID) between search model and target X-ray crystal structure. Solutions derived from X-ray crystal structure search models are colored red, and solutions derived from ‘fc’-trimmed NMR structure ensemble search models are colored blue. (B) Plot of free R factor values of final ARP/wARP models vs sequence identity between search models and target X-ray structures. Solutions derived from X-ray crystal structure search models are colored red, and solutions derived from ‘fc’-trimmed NMR structure ensemble search models are colored blue. See also Tables S6 and S7.
Similar articles
- Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Ramelot TA, Raman S, Kuzin AP, Xiao R, Ma LC, Acton TB, Hunt JF, Montelione GT, Baker D, Kennedy MA. Ramelot TA, et al. Proteins. 2009 Apr;75(1):147-67. doi: 10.1002/prot.22229. Proteins. 2009. PMID: 18816799 Free PMC article. - Protein NMR structures refined with Rosetta have higher accuracy relative to corresponding X-ray crystal structures.
Mao B, Tejero R, Baker D, Montelione GT. Mao B, et al. J Am Chem Soc. 2014 Feb 5;136(5):1893-906. doi: 10.1021/ja409845w. Epub 2014 Jan 23. J Am Chem Soc. 2014. PMID: 24392845 Free PMC article. - Crystallographic phasing with NMR models: an envelope approach.
Zhang W, Zhang T, Zhang H, Hao Q. Zhang W, et al. Acta Crystallogr D Biol Crystallogr. 2014 Jul;70(Pt 7):1977-82. doi: 10.1107/S1399004714009754. Epub 2014 Jun 29. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 25004974 - Rosetta Structure Prediction as a Tool for Solving Difficult Molecular Replacement Problems.
DiMaio F. DiMaio F. Methods Mol Biol. 2017;1607:455-466. doi: 10.1007/978-1-4939-7000-1_19. Methods Mol Biol. 2017. PMID: 28573585 Review. - Combination of NMR spectroscopy and X-ray crystallography offers unique advantages for elucidation of the structural basis of protein complex assembly.
Feng W, Pan L, Zhang M. Feng W, et al. Sci China Life Sci. 2011 Feb;54(2):101-11. doi: 10.1007/s11427-011-4137-2. Epub 2011 Feb 14. Sci China Life Sci. 2011. PMID: 21318479 Review.
Cited by
- Critical assessment of methods of protein structure prediction: Progress and new directions in round XI.
Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Moult J, et al. Proteins. 2016 Sep;84 Suppl 1(Suppl 1):4-14. doi: 10.1002/prot.25064. Epub 2016 Jun 1. Proteins. 2016. PMID: 27171127 Free PMC article. - Automatic 13C chemical shift reference correction for unassigned protein NMR spectra.
Chen X, Smelter A, Moseley HNB. Chen X, et al. J Biomol NMR. 2018 Oct;72(1-2):11-28. doi: 10.1007/s10858-018-0202-5. Epub 2018 Aug 10. J Biomol NMR. 2018. PMID: 30097912 Free PMC article. - Quality assessment of protein NMR structures.
Rosato A, Tejero R, Montelione GT. Rosato A, et al. Curr Opin Struct Biol. 2013 Oct;23(5):715-24. doi: 10.1016/j.sbi.2013.08.005. Epub 2013 Sep 21. Curr Opin Struct Biol. 2013. PMID: 24060334 Free PMC article. Review. - Application of the AMPLE cluster-and-truncate approach to NMR structures for molecular replacement.
Bibby J, Keegan RM, Mayans O, Winn MD, Rigden DJ. Bibby J, et al. Acta Crystallogr D Biol Crystallogr. 2013 Nov;69(Pt 11):2194-201. doi: 10.1107/S0907444913018453. Epub 2013 Oct 12. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24189230 Free PMC article. - The expanded FindCore method for identification of a core atom set for assessment of protein structure prediction.
Snyder DA, Grullon J, Huang YJ, Tejero R, Montelione GT. Snyder DA, et al. Proteins. 2014 Feb;82 Suppl 2(0 2):219-30. doi: 10.1002/prot.24490. Proteins. 2014. PMID: 24327305 Free PMC article.
References
- Anderson DH, Weiss MS, Eisenberg D. A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi. Acta Crystallogr D Biol Crystallogr. 1996;52:469–480. - PubMed
- Aramini JM, Ma L, Lee H, Zhao L, Cunningham K, Ciccosanti C, Janjua H, Fang Y, Xiao R, Krug RM, Montelione GT. Solution NMR structure of the monomeric W187R mutant of A/Udorn NS1 effector domain. Northeast Structural Genomics target OR8C[W187R] journal. 2009 page numbers, etc.
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM095693/GM/NIGMS NIH HHS/United States
- U54 GM074958/GM/NIGMS NIH HHS/United States
- U54 GM094597/GM/NIGMS NIH HHS/United States
- U54 GM094597-01/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources