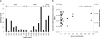Comparison of innovative molecular approaches and standard spore assays for assessment of surface cleanliness - PubMed (original) (raw)
Comparative Study
. 2011 Aug;77(15):5438-44.
doi: 10.1128/AEM.00192-11. Epub 2011 Jun 7.
Affiliations
- PMID: 21652744
- PMCID: PMC3147454
- DOI: 10.1128/AEM.00192-11
Comparative Study
Comparison of innovative molecular approaches and standard spore assays for assessment of surface cleanliness
Moogega Cooper et al. Appl Environ Microbiol. 2011 Aug.
Abstract
A bacterial spore assay and a molecular DNA microarray method were compared for their ability to assess relative cleanliness in the context of bacterial abundance and diversity on spacecraft surfaces. Colony counts derived from the NASA standard spore assay were extremely low for spacecraft surfaces. However, the PhyloChip generation 3 (G3) DNA microarray resolved the genetic signatures of a highly diverse suite of microorganisms in the very same sample set. Samples completely devoid of cultivable spores were shown to harbor the DNA of more than 100 distinct microbial phylotypes. Furthermore, samples with higher numbers of cultivable spores did not necessarily give rise to a greater microbial diversity upon analysis with the DNA microarray. The findings of this study clearly demonstrated that there is not a statistically significant correlation between the cultivable spore counts obtained from a sample and the degree of bacterial diversity present. Based on these results, it can be stated that validated state-of-the-art molecular techniques, such as DNA microarrays, can be utilized in parallel with classical culture-based methods to further describe the cleanliness of spacecraft surfaces.
Figures
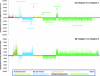
Fig. 1.
PhyloChip analysis of hybridization intensity differences between the three spore abundance categories. (a) Hybridization intensity difference of category B (1 to 50 spores) compared to category A (0 spores). (b) Hybridization intensity difference of category C (>50 spores) compared to category B. Bars above the zero line represent bacteria that increased in abundance relative to category A and category B bacteria for panels a and b, respectively; bars below the zero line represent those bacterial DNAs that declined in abundance.
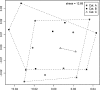
Fig. 2.
NMDS analysis based on log2 transformation·1 × 103 hybridization intensity of OTUs reveals that the spore categorical groupings are from similar community structures, as they greatly overlap. x axis, dimension 1; y axis, dimension 2.
Fig. 3.
(a) Comparison of subfamily richness and spore counts grouped by the spore abundance categories and based on the NASA standard spore assay. The results show that there is no strong correlation between sample category and subfamily count. (b) Calculated cell density based on 16S rRNA gene copies compared to spore counts and total hybridization intensity. The average number of 16S rRNA gene copies is 3.6 per cell (17, 22, 32). Gene copies correlate similarly to hybridization intensities but show no correlation to spore counts.
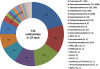
Fig. 4.
Overall microbial diversity grouped by taxa. Numbers of subfamilies and OTUs detected per taxon are given in the parentheses. Percentages lower than 2% are not displayed.
Similar articles
- Clean room microbiome complexity impacts planetary protection bioburden.
Hendrickson R, Urbaniak C, Minich JJ, Aronson HS, Martino C, Stepanauskas R, Knight R, Venkateswaran K. Hendrickson R, et al. Microbiome. 2021 Dec 4;9(1):238. doi: 10.1186/s40168-021-01159-x. Microbiome. 2021. PMID: 34861887 Free PMC article. - Microbial monitoring of spacecraft and associated environments.
La Duc MT, Kern R, Venkateswaran K. La Duc MT, et al. Microb Ecol. 2004 Feb;47(2):150-8. doi: 10.1007/s00248-003-1012-0. Epub 2004 Feb 2. Microb Ecol. 2004. PMID: 14749906 - Recovery of bacillus spore contaminants from rough surfaces: a challenge to space mission cleanliness control.
Probst A, Facius R, Wirth R, Wolf M, Moissl-Eichinger C. Probst A, et al. Appl Environ Microbiol. 2011 Mar;77(5):1628-37. doi: 10.1128/AEM.02037-10. Epub 2011 Jan 7. Appl Environ Microbiol. 2011. PMID: 21216908 Free PMC article. - Detecting the dormant: a review of recent advances in molecular techniques for assessing the viability of bacterial endospores.
Mohapatra BR, La Duc MT. Mohapatra BR, et al. Appl Microbiol Biotechnol. 2013 Sep;97(18):7963-75. doi: 10.1007/s00253-013-5115-3. Epub 2013 Aug 3. Appl Microbiol Biotechnol. 2013. PMID: 23912118 Review. - High-throughput methods for analysis of the human oral microbiome.
Pozhitkov AE, Beikler T, Flemmig T, Noble PA. Pozhitkov AE, et al. Periodontol 2000. 2011 Feb;55(1):70-86. doi: 10.1111/j.1600-0757.2010.00380.x. Periodontol 2000. 2011. PMID: 21134229 Review. No abstract available.
Cited by
- Resistance of bacterial endospores to outer space for planetary protection purposes--experiment PROTECT of the EXPOSE-E mission.
Horneck G, Moeller R, Cadet J, Douki T, Mancinelli RL, Nicholson WL, Panitz C, Rabbow E, Rettberg P, Spry A, Stackebrandt E, Vaishampayan P, Venkateswaran KJ. Horneck G, et al. Astrobiology. 2012 May;12(5):445-56. doi: 10.1089/ast.2011.0737. Astrobiology. 2012. PMID: 22680691 Free PMC article. - New perspectives on microbial community distortion after whole-genome amplification.
Probst AJ, Weinmaier T, DeSantis TZ, Santo Domingo JW, Ashbolt N. Probst AJ, et al. PLoS One. 2015 May 26;10(5):e0124158. doi: 10.1371/journal.pone.0124158. eCollection 2015. PLoS One. 2015. PMID: 26010362 Free PMC article. - Metagenomic Methods for Addressing NASA's Planetary Protection Policy Requirements on Future Missions: A Workshop Report.
Green SJ, Torok T, Allen JE, Eloe-Fadrosh E, Jackson SA, Jiang SC, Levine SS, Levy S, Schriml LM, Thomas WK, Wood JM, Tighe SW. Green SJ, et al. Astrobiology. 2023 Aug;23(8):897-907. doi: 10.1089/ast.2022.0044. Epub 2023 Apr 26. Astrobiology. 2023. PMID: 37102710 Free PMC article. - New perspectives on viable microbial communities in low-biomass cleanroom environments.
Vaishampayan P, Probst AJ, La Duc MT, Bargoma E, Benardini JN, Andersen GL, Venkateswaran K. Vaishampayan P, et al. ISME J. 2013 Feb;7(2):312-24. doi: 10.1038/ismej.2012.114. Epub 2012 Oct 11. ISME J. 2013. PMID: 23051695 Free PMC article. - Clean room microbiome complexity impacts planetary protection bioburden.
Hendrickson R, Urbaniak C, Minich JJ, Aronson HS, Martino C, Stepanauskas R, Knight R, Venkateswaran K. Hendrickson R, et al. Microbiome. 2021 Dec 4;9(1):238. doi: 10.1186/s40168-021-01159-x. Microbiome. 2021. PMID: 34861887 Free PMC article.
References
- ASTM 2009. ASTM E2362-09. Standard practice for evaluation of pre-saturated or impregnated towelettes for hard surface disinfection. ASTM book of standards, vol. 11.05, p. 1622 ASTM International, West Conshohocken, PA
- Chivian D., et al. 2008. Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322:275–278 - PubMed
- Committee on Preventing the Forward Contamination of Mars, National Research Council 2006. Preventing the forward contamination of Mars. The National Academies Press, Washington, DC
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources