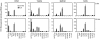Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype - PubMed (original) (raw)
Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype
Paul C Norris et al. J Leukoc Biol. 2011 Sep.
Abstract
Eicosanoid metabolism differs in profile and quantity between macrophages of different tissue origin and method of elicitation, as well as between primary and immortalized macrophages after activation with inflammatory stimuli. Using a lipidomic approach, we comprehensively analyzed the eicosanoids made by murine RPMs, TGEMs, BMDM, and the macrophage-like cell line RAW after stimulation with the TLR-4-specific agonist KLA. Direct correlation among total COX metabolites, COX side-products (11-HETE, 15-HETE), COX-2 mRNA, and protein at 8 h was found when comparing each cell type. Comprehensive qPCR analysis was used to compare relative transcript levels between the terminal prostanoid synthases themselves as well as between each cell type. Levels of PGE(2), PGD(2), and TxB(2) generally correlated with enzyme transcript expression of PGES, PGDS, and TBXS, providing evidence of comparable enzyme activities. PGIS transcript was expressed only in RPM and TGEM macrophages and at an exceptionally low level, despite high metabolite production compared with other synthases. Presence of PGIS in RPM and TGEM also lowered the production of PGE(2) versus PGD(2) by approximately tenfold relative to BMDM and RAW cells, which lacked this enzyme. Our results demonstrate that delayed PG production depends on the maximal level of COX-2 expression in different macrophages after TLR-4 stimulation. Also, the same enzymes in each cell largely dictate the profile of eicosanoids produced depending on the ratios of expression between them, with the exception of PGIS, which appears to have much greater synthetic capacity and competes selectively with mPGES-1.
Figures
Figure 1.. Lipidomic analysis of RPM, TGEM, BMDM, and RAW macrophages.
Heat map representing fold-change in the extracellular medium levels of 140 eicosanoid species after stimulation with the TLR-4-specific receptor agonist KLA (100 ng/ml) relative to unstimulated PBS control at 0, 8, and 24 h time-points. Increases in metabolite levels are indicated by red, decreases by green, and detectable but unchanged levels by gray. Metabolites below the limit of detection are indicated by black; n = 3 individual biological replicates/time-point/group. PGDH, PG dehydrogenase; β-ox, β-oxidation; LTAH, LTA4 hydrolase; LTCS, LTC4 synthase; HEDH, HETE dehydrogenase; sEH, soluble epoxide hydrolase.
Figure 2.. Quantitative eicosanoid production profiles of different TLR-4 agonist-stimulated macrophages.
Different macrophage types incubated in the absence (open bars) and presence (shaded bars) of KLA (100 ng/ml) from the same experiment as Fig. 1. Extracellular medium was removed at 8 h (upper row) and 24 h (lower row) and was analyzed for eicosanoid levels by MS. The data are expressed as mean values ±
sem
of three biological replicates.
Figure 3.. COX-2 activity and expression.
Comparison of 8 h (A) total arachidonate COX-derived eicosanoids, (B) 11-HETE, and (C) 15-HETE from the same experiment as in Fig. 1; relative expression level of (D) COX-2 mRNA and (E) COX-1 mRNA at 0, 8, and 24 h; and (F) 8 h COX-2 protein in RPM, TGEM, BMDM, and RAW macrophages after KLA (100 ng/ml) stimulation. The data are expressed as mean values ±
sem
of three biological replicates. WB, Western blot.
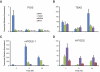
Figure 4.. Expression of terminal prostanoid synthases.
Comparison of transcript expression of (A) PGIS, (B) TBXS (TBXAS), (C) mPGES-1, and (D) H-PGDS synthases after 0, 8, and 24 h of KLA (100 ng/ml) stimulation in RPM (blue), TGEM (green), BMDM (purple), and RAW (red) macrophages. The data are expressed as mean values ±
sem
of three biological replicates.
Figure 5.. Correlation between PGE2:PGD2 ratio and respective enzyme transcript expression ratio.
(A) Eicosanoid ratios of PGE2:PGD2 after 8 h and 24 h KLA stimulation, where PGD2, PGJ2, 15d PGD2, and 15d PGJ2 were summed. (B) Transcript expression ratios of mPGES-1:H-PGDS using average expression of 0 h and 8 h time-points (0–8 h) and 0, 8, and 24 h time-points (0–24) in RPM (blue), TGEM (green), BMDM (purple), and RAW (red) macrophages stimulated with KLA (100 ng/ml). The data are expressed as mean values ±
sem
of three biological replicates.
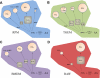
Figure 6.. Overview of prostanoid synthase expression and relative activity in different macrophage cell types after long-term TLR-4 activation.
Metabolism of AA in (A) RPM, (B) TGEM, (C) BMDM, and (D) RAW through induced COX-2 leads to a pool of PGH2 that is metabolized by TBXS, PGIS, PGDS (H-PGDS), and induced PGES (mPGES-1). Percentages of metabolites (circles) and enzyme transcripts (squares) in each cell type are represented by area after 8 h KLA stimulation. Metabolites and expressed levels of PGES, PGDS, and TBXS are roughly proportional, although the presence of PGIS appears to selectively draw substrate away from PGES but not PGDS. Low constitutive expression level of PGIS in RPM and TGEM macrophages with disproportionately high levels of PGI2 suggests a significantly higher synthetic rate compared with other prostanoid synthases.
Similar articles
- Lipopolysaccharide-induced expression of microsomal prostaglandin E synthase-1 mediates late-phase PGE2 production in bone marrow derived macrophages.
Xiao L, Ornatowska M, Zhao G, Cao H, Yu R, Deng J, Li Y, Zhao Q, Sadikot RT, Christman JW. Xiao L, et al. PLoS One. 2012;7(11):e50244. doi: 10.1371/journal.pone.0050244. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226252 Free PMC article. - Modeling of eicosanoid fluxes reveals functional coupling between cyclooxygenases and terminal synthases.
Kihara Y, Gupta S, Maurya MR, Armando A, Shah I, Quehenberger O, Glass CK, Dennis EA, Subramaniam S. Kihara Y, et al. Biophys J. 2014 Feb 18;106(4):966-75. doi: 10.1016/j.bpj.2014.01.015. Biophys J. 2014. PMID: 24559999 Free PMC article. - Redirection of eicosanoid metabolism in mPGES-1-deficient macrophages.
Trebino CE, Eskra JD, Wachtmann TS, Perez JR, Carty TJ, Audoly LP. Trebino CE, et al. J Biol Chem. 2005 Apr 29;280(17):16579-85. doi: 10.1074/jbc.M412075200. Epub 2005 Feb 18. J Biol Chem. 2005. PMID: 15722356 - Prostaglandin E synthases: Understanding their pathophysiological roles through mouse genetic models.
Hara S, Kamei D, Sasaki Y, Tanemoto A, Nakatani Y, Murakami M. Hara S, et al. Biochimie. 2010 Jun;92(6):651-9. doi: 10.1016/j.biochi.2010.02.007. Epub 2010 Feb 14. Biochimie. 2010. PMID: 20159030 Review. - Membrane prostaglandin E synthase-1: a novel therapeutic target.
Samuelsson B, Morgenstern R, Jakobsson PJ. Samuelsson B, et al. Pharmacol Rev. 2007 Sep;59(3):207-24. doi: 10.1124/pr.59.3.1. Pharmacol Rev. 2007. PMID: 17878511 Review.
Cited by
- Macrophage energy metabolism in cardiometabolic disease.
Wong A, Sun Q, Latif II, Karwi QG. Wong A, et al. Mol Cell Biochem. 2024 Aug 29. doi: 10.1007/s11010-024-05099-6. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39198360 - Infection, Inflammation, and Immunity in Sepsis.
Das UN. Das UN. Biomolecules. 2023 Aug 31;13(9):1332. doi: 10.3390/biom13091332. Biomolecules. 2023. PMID: 37759732 Free PMC article. Review. - Polyunsaturated and Saturated Oxylipin Plasma Levels Allow Monitoring the Non-Alcoholic Fatty Liver Disease Progression to Severe Stages.
Ferrer MD, Reynés C, Monserrat-Mesquida M, Quetglas-Llabrés M, Bouzas C, García S, Mateos D, Casares M, Gómez C, Ugarriza L, Tur JA, Sureda A, Pons A. Ferrer MD, et al. Antioxidants (Basel). 2023 Mar 13;12(3):711. doi: 10.3390/antiox12030711. Antioxidants (Basel). 2023. PMID: 36978959 Free PMC article. - Blockage of lamin-A/C loss diminishes the pro-inflammatory macrophage response.
Mehl JL, Earle A, Lammerding J, Mhlanga M, Vogel V, Jain N. Mehl JL, et al. iScience. 2022 Nov 7;25(12):105528. doi: 10.1016/j.isci.2022.105528. eCollection 2022 Dec 22. iScience. 2022. PMID: 36465100 Free PMC article. - Biosynthesis of prostaglandin 15dPGJ2 -glutathione and 15dPGJ2-cysteine conjugates in macrophages and mast cells via MGST3.
Steinmetz-Späh J, Liu J, Singh R, Ekoff M, Boddul S, Tang X, Bergqvist F, Idborg H, Heitel P, Rönnberg E, Merk D, Wermeling F, Haeggström JZ, Nilsson G, Steinhilber D, Larsson K, Korotkova M, Jakobsson PJ. Steinmetz-Späh J, et al. J Lipid Res. 2022 Dec;63(12):100310. doi: 10.1016/j.jlr.2022.100310. Epub 2022 Nov 9. J Lipid Res. 2022. PMID: 36370807 Free PMC article.
References
- Six D. A., Dennis E. A. (2000) The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta 1488, 1–19 - PubMed
- Schaloske R. H., Dennis E. A. (2006) The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259 - PubMed
- Funk C. D. (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 - PubMed
- Simmons D. L., Botting R. M., Hla T. (2004) Cyclooxygenase isozymes: the biology of prostaglandin synthesis and inhibition. Pharmacol. Rev. 56, 387–437 - PubMed
- Smith W. L., DeWitt D. L., Garavito R. M. (2000) Cyclooxygenases: structural, cellular, and molecular biology. Annu. Rev. Biochem. 69, 145–182 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM069338/GM/NIGMS NIH HHS/United States
- T32 GM007752./GM/NIGMS NIH HHS/United States
- R01 GM064611/GM/NIGMS NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- R01 GM64611/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous