PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria - PubMed (original) (raw)
. 2011 Sep 1;39(17):7750-63.
doi: 10.1093/nar/gkr470. Epub 2011 Jun 11.
Affiliations
- PMID: 21666256
- PMCID: PMC3177208
- DOI: 10.1093/nar/gkr470
PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria
Joanna Rorbach et al. Nucleic Acids Res. 2011.
Abstract
Polyadenylation of mRNA in human mitochondria is crucial for gene expression and perturbation of poly(A) tail length has been linked to a human neurodegenerative disease. Here we show that 2'-phosphodiesterase (2'-PDE), (hereafter PDE12), is a mitochondrial protein that specifically removes poly(A) extensions from mitochondrial mRNAs both in vitro and in mitochondria of cultured cells. In eukaryotes, poly(A) tails generally stabilize mature mRNAs, whereas in bacteria they increase mRNA turnover. In human mitochondria, the effects of increased PDE12 expression were transcript dependent. An excess of PDE12 led to an increase in the level of three mt-mRNAs (ND1, ND2 and CytB) and two (CO1 and CO2) were less abundant than in mitochondria of control cells and there was no appreciable effect on the steady-state level of the remainder of the mitochondrial transcripts. The alterations in poly(A) tail length accompanying elevated PDE12 expression were associated with severe inhibition of mitochondrial protein synthesis, and consequently respiratory incompetence. Therefore, we propose that mRNA poly(A) tails are important in regulating protein synthesis in human mitochondria, as it is the case for nuclear-encoded eukaryotic mRNA.
Figures
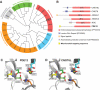
Figure 1.
Computational analysis of PDE12. (A) The unrooted classification tree of human EEP proteins. The sequences of the catalytic domain of all human proteins of the EEP family (PF03372) were extracted from the Pfam database (see
Supplementary Table S1
for details) and aligned using ClustalW2. Five major subgroups identified are indicated on the tree: inositol phosphatases (orange), Sphingomyelin phosphodiesterases (grey), Endo- and exonucleases (green), DNase I-like proteins (blue) and Deadenylases (red). (B) Domain architecture of confirmed and putative human deadenylases of the EEP family. Functional domains of human proteins of the ‘Deadenylases’ subgroup identified in (A) are schematically represented. The position of the EEP domain is represented in each protein by a red box containing the percentage of identity between this domain and that of PDE12. (C and D) Comparison of the active site structure of PDE12 and other EEP deadenylases. The active site structural motif of PDE12 (C) predicted based on the known crystal structure of the homologous EEP deadenylase CNOT6L (D) using the 3D-Jury algorithm (42) was modelled using the MODELLER 9v1 software (43).
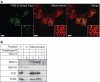
Figure 2.
Mitochondrial localization of PDE12. (A) The intra-cellular localization of PDE12 by immunofluorescence. The cDNA encoding the Strep2- and Flag-tagged variant of PDE12 was transiently transfected into HOS cells, the protein product was detected by anti-Flag antibody and visualized by secondary antibodies conjugated with FITC (green). Mitochondria were stained with MitoTracker Red CMXRos (red). Co-localization of the green and red signal appears yellow on digitally overlaid images (Merge). (B) Location of PDE12 in subcellular fractions. The HOS cells were fractionated into cytosol (C, lane 2) and mitochondria (lanes 3–5) as described ‘Material and Methods’ section. The mitochondrial fraction was treated with 25 µg/ml proteinase K in the absence (lane 4) or presence of 1% Triton X-100 (lane 5). ‘T’ denotes the total cell lysate. The protein fractions were analysed by western blotting using antibodies to endogenous PDE12. The location of PDE12 was compared with that of the following marker proteins: TFAM (mitochondrial matrix), CO2 (mitochondrial inner membrane) and GAPDH (cytosol).
Figure 3.
Enzymatic activity of PDE12. (A) 3′→5′ exoribonucleolytic activity of PDE12. 8.5 pmoles of PDE12.Strep2.Flag or the E351A catalytic mutant purified from human mitochondria were incubated with 5 pmols of radioactively labelled 25 nt-long homopolymeric adenosine RNA (A25), 25 nt-long homopolymeric adenosine DNA (dA25), 25 nt-long homopolymeric adenosine RNA labelled on the 3′-end with pCp (A25pCp) or 25 nt-long homopolymeric adenosine RNA with cytosine ribonucleotide at the 3′-end (A25pC-OH) for the indicated time. The products were separated on a 10% urea polyacrylamide gel and subjected to autoradiography. (B) Sequence specificity of PDE12. 8.5 pmoles of PDE12.Strep2.Flag or the E351A catalytic mutant were incubated with 5 pmoles of radioactively labelled 25 nt-long homopolymeric RNAs consisting of adenine (A25), cytosine (C25), guanine (G25) or uridine (U25) nucleotides for the indicated time. The products were separated on a 10% urea polyacrylamide gel and subjected to autoradiography. (C) Deadenylase activity of PDE12 on synthetic mt-RNA substrates. About 8.5 pmols of PDE12.Strep2.Flag or the E351A catalytic mutant were incubated with synthetic transcripts produced by run-off T7 transcription in the presence of radioactive UTP. The following RNAs were tested: 111 nt-long 3′-region of the ND1 ORF (3′ND1, lanes 1–3), 111 nt-long 3′-part of the ND1 ORF containing a 50 nt poly(A) tail (3′ND1-A50, lanes 4–6), 111-nt long 3′-part of the CO2 ORF (3′CO2, lanes 7–9) or 111 nt-long 3′-part of the CO2 ORF containing a 50-nt poly(A) tail (3′CO2-A50, lanes 10–12). The products were separated on a 5% urea polyacrylamide gel and subjected to autoradiography.
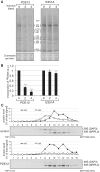
Figure 4.
Mitochondrial transcripts in cells overexpressing PDE12. (A) Inducible expression of PDE12 and the E351A catalytic mutant in HEK293T cells. Western blot illustrating a time course of expression of PDE12.Strep2.Flag or the E351A mutant induced with 50 ng/ml doxycycline. The PDE12 protein was detected using anti-Flag antibody; anti-VDAC-1 antibodies were used as a mitochondrial loading control. (B) Analysis of the poly(A) tail length of mitochondrial transcripts upon PDE12 induction. Mitochondrial ND1 or CO2 mRNAs were analysed by cRT–PCR and sequencing as described in ‘Materials and Methods’ section. Each data point represents a single clone analysed for the HEK293T parental cells (triangle), wild-type PDE12 (circle) and the E351A mutant (square). The length of poly(A) extensions are given on the _Y_-axis (note that the values below zero indicate transcripts that were truncated beyond the processing/polyadenylation site). Doxycycline induction times are given on the _X_-axis. The horizontal lines represent average lengths of poly(A) tails. (C) Northern blots of mitochondrial transcripts upon overexpression of PDE12. Total RNA from parental HEK293T cells (H), uninduced cells (0 days) and cells expressing PDE12 for 2 or 4 days (induced with 50 ng/ml doxycycline) from two independent experiments (‘expt. 1′ and ‘expt. 2′) were analysed by northern blots using probes specific for all mitochondrial transcripts. Nuclear-encoded 28 S rRNA was used as a loading control. (D) Quantification of steady-state levels of mitochondrial transcripts in cells overexpressing PDE12. The values of the relative RNA levels (mt-RNA/28 S rRNA) were obtained by quantifying PhosphorImager scans of northern blots using ImageQuant software and normalized for the values obtained from uninduced cells. *P < 0.05, **P < 0.01, ***P < 0.001; n = 3, Error bars = 1 SD. (E) mtDNA copy number in cells overexpressing PDE12. Comparative Q-PCR of the mitochondrial CytB gene and single copy nuclear gene (APP) was used with total DNA isolated from cells overexpressing PDE12 for the indicated time. *P < 0.05, **P < 0.01, ***P < 0.001; n = 3, Error bars = 1 SD.
Figure 5.
Respiratory chain function upon PDE12 overexpression. (A) Growth of cells overexpressing PDE12 on galactose and glucose media. Growth curves of parental HEK293 cells or transfectants-expressing PDE12 or the E351A mutant in galactose or glucose (inset) media and induced for the indicated time. (B) Mitochondrial membrane potential in cells overexpressing PDE12. Mitochondrial membrane potential was assessed by a quantitative FACS analysis of TMRE intensity. Twenty thousand cells stained with TMRE were analysed for each sample. The percentages of TMRE intensity were normalized to that of HEK293T cells (=100%). The mtDNA-less HEK293T cells were used as a negative control. **P < 0.01, ***P < 0.001; two-tailed Student’s _t_-test; n = 3, Error bars = 1 SD. (C) Steady-state level of the OXPHOS subunits in cells overexpressing PDE12. Steady-state protein level of subunits of respiratory chain complexes in uninduced control cells or cells overexpressing PDE12 or its catalytic mutant (E351A) for the indicated time were analysed by western blot. β-Actin was used as a loading control. (D) OCR in cells overexpressing PDE12. OCR measured in an extracellular flux Seahorse instrument in a quadruplicate population of control uninduced or cells overexpressing PDE12 or its catalytic mutant (E351A) for 4 days. The wells containing cells were sequentially injected with 20 mM 2-DG to inhibit glycolysis, 100 nM oligomycin to inhibit ATP synthase, 1 µM FCCP to uncouple the respiratory chain and 200 nM rotenone to inhibit complex I.** P < 0.01; two-tailed Student’s _t_-test; n = 3, Error bars = 1 SD.
Figure 6.
Mitochondrial translation in cells overexpressing PDE12**.** (A) Products of mitochondrial translation were labelled with 35S 36methionine for 15 min after expression of PDE12 or the E351A catalytic mutant in HEK293T cells for 2 or 4 days as described in ‘Materials and Methods’ section. Mitochondrial proteins were separated by a 4–12% gradient SDS–PAGE and visualized by autoradiography. To validate equal protein loading, a small section of the gel was stained with Coomassie. (B) Radiolabelled products of mitochondrial translation as in (A) were quantified using ImageQuant software following exposure to a PhosphorImager cassette. ***P < 0.001; two-tailed Student’s _t_-test; n = 4, Error bars = 1 SD. (C) Mitochondrial ribosome profile in cells overexpressing PDE12. Cell lysates of HEK293T cells overexpressing PDE12 for 4 days or control cells were separated on a 10–30% (v:v) isokinetic sucrose gradient. Fractions obtained for control and PDE12 overexpressing cells were analysed by western blot simultaneously with antibodies to MRPL3 (39 S mitoribosomal subunit) and DAP3 (28 S mitoribosomal subunit).
Similar articles
- Maturation of selected human mitochondrial tRNAs requires deadenylation.
Pearce SF, Rorbach J, Van Haute L, D'Souza AR, Rebelo-Guiomar P, Powell CA, Brierley I, Firth AE, Minczuk M. Pearce SF, et al. Elife. 2017 Jul 26;6:e27596. doi: 10.7554/eLife.27596. Elife. 2017. PMID: 28745585 Free PMC article. - Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition.
Wydro M, Bobrowicz A, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZM. Wydro M, et al. Nucleic Acids Res. 2010 Jun;38(11):3732-42. doi: 10.1093/nar/gkq068. Epub 2010 Feb 9. Nucleic Acids Res. 2010. PMID: 20144953 Free PMC article. - Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA.
Slomovic S, Schuster G. Slomovic S, et al. RNA. 2008 Feb;14(2):310-23. doi: 10.1261/rna.697308. Epub 2007 Dec 14. RNA. 2008. PMID: 18083837 Free PMC article. - PARN-like Proteins Regulate Gene Expression in Land Plant Mitochondria by Modulating mRNA Polyadenylation.
Hirayama T. Hirayama T. Int J Mol Sci. 2021 Oct 5;22(19):10776. doi: 10.3390/ijms221910776. Int J Mol Sci. 2021. PMID: 34639116 Free PMC article. Review. - RNA degradation in human mitochondria: the journey is not finished.
Santonoceto G, Jurkiewicz A, Szczesny RJ. Santonoceto G, et al. Hum Mol Genet. 2024 May 22;33(R1):R26-R33. doi: 10.1093/hmg/ddae043. Hum Mol Genet. 2024. PMID: 38779774 Free PMC article. Review.
Cited by
- Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci.
Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, Szczesny RJ. Borowski LS, et al. Nucleic Acids Res. 2013 Jan;41(2):1223-40. doi: 10.1093/nar/gks1130. Epub 2012 Dec 5. Nucleic Acids Res. 2013. PMID: 23221631 Free PMC article. - Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels.
Szczesny RJ, Hejnowicz MS, Steczkiewicz K, Muszewska A, Borowski LS, Ginalski K, Dziembowski A. Szczesny RJ, et al. Nucleic Acids Res. 2013 Mar 1;41(5):3144-61. doi: 10.1093/nar/gkt029. Epub 2013 Jan 28. Nucleic Acids Res. 2013. PMID: 23358826 Free PMC article. - Mechanisms and regulation of protein synthesis in mitochondria.
Kummer E, Ban N. Kummer E, et al. Nat Rev Mol Cell Biol. 2021 May;22(5):307-325. doi: 10.1038/s41580-021-00332-2. Epub 2021 Feb 16. Nat Rev Mol Cell Biol. 2021. PMID: 33594280 Review. - Mitochondrial poly(A) polymerase and polyadenylation.
Chang JH, Tong L. Chang JH, et al. Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):992-7. doi: 10.1016/j.bbagrm.2011.10.012. Epub 2011 Dec 7. Biochim Biophys Acta. 2012. PMID: 22172994 Free PMC article. Review. - A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression.
Wilson WC, Hornig-Do HT, Bruni F, Chang JH, Jourdain AA, Martinou JC, Falkenberg M, Spåhr H, Larsson NG, Lewis RJ, Hewitt L, Baslé A, Cross HE, Tong L, Lebel RR, Crosby AH, Chrzanowska-Lightowlers ZM, Lightowlers RN. Wilson WC, et al. Hum Mol Genet. 2014 Dec 1;23(23):6345-55. doi: 10.1093/hmg/ddu352. Epub 2014 Jul 9. Hum Mol Genet. 2014. PMID: 25008111 Free PMC article.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases