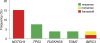Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation - PubMed (original) (raw)
. 2011 Jul 4;208(7):1389-401.
doi: 10.1084/jem.20110921. Epub 2011 Jun 13.
Silvia Rasi, Davide Rossi, Vladimir Trifonov, Hossein Khiabanian, Jing Ma, Adina Grunn, Marco Fangazio, Daniela Capello, Sara Monti, Stefania Cresta, Ernesto Gargiulo, Francesco Forconi, Anna Guarini, Luca Arcaini, Marco Paulli, Luca Laurenti, Luigi M Larocca, Roberto Marasca, Valter Gattei, David Oscier, Francesco Bertoni, Charles G Mullighan, Robin Foá, Laura Pasqualucci, Raul Rabadan, Riccardo Dalla-Favera, Gianluca Gaidano
Affiliations
- PMID: 21670202
- PMCID: PMC3135373
- DOI: 10.1084/jem.20110921
Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation
Giulia Fabbri et al. J Exp Med. 2011.
Abstract
The pathogenesis of chronic lymphocytic leukemia (CLL), the most common leukemia in adults, is still largely unknown. The full spectrum of genetic lesions that are present in the CLL genome, and therefore the number and identity of dysregulated cellular pathways, have not been identified. By combining next-generation sequencing and copy number analysis, we show here that the typical CLL coding genome contains <20 clonally represented gene alterations/case, including predominantly nonsilent mutations, and fewer copy number aberrations. These analyses led to the discovery of several genes not previously known to be altered in CLL. Although most of these genes were affected at low frequency in an expanded CLL screening cohort, mutational activation of NOTCH1, observed in 8.3% of CLL at diagnosis, was detected at significantly higher frequency during disease progression toward Richter transformation (31.0%), as well as in chemorefractory CLL (20.8%). Consistent with the association of NOTCH1 mutations with clinically aggressive forms of the disease, NOTCH1 activation at CLL diagnosis emerged as an independent predictor of poor survival. These results provide initial data on the complexity of the CLL coding genome and identify a dysregulated pathway of diagnostic and therapeutic relevance.
Figures
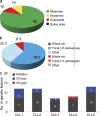
Figure 1.
Overall load of genetic lesions in CLL. (a) Frequency of Sanger-validated somatic nonsilent mutations identified through WES in the five CLL discovery cases. (b) Overall frequency of somatically acquired CNA according to aberration type in the 5 CLL discovery cases. (c) Combined load of somatically acquired nonsilent mutations and CNA identified in the CLL discovery panel. CN, copy number.
Figure 2.
Recurrent mutations in CLL. Percentage of CLL primary cases harboring mutations in candidate genes after targeted resequencing of an expanded screening dataset (n = 48 CLL cases). The final number of mutated cases over total analyzed (including the 5 discovery CLL) for each gene is as follows: NOTCH1, 8/53; TP53, 4/53; PLEKHG5, 2/53; TGM7, 2/53; BIRC3, 2/53.
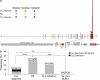
Figure 3.
NOTCH1 is mutated in a large fraction of RS cases and chemorefractory CLL. (a) Schematic diagram of the NOTCH1 gene (top) and protein (bottom), with its conserved functional domains (EGF-like repeats; LNR, LIN-12/NOTCH repeats; HD, heterodimerization; TM, transmembrane; Ank, Ankyrin repeats; TAD, transactivation; PEST). Color-coded shapes indicate the position of the mutations found in CLL at diagnosis, RS, and chemorefractory CLL. (b) Prevalence of NOTCH1 mutations in CLL at diagnosis, RS, chemorefractory CLL, and de novo DLBCL; numbers on top indicate the actual number of mutated samples over total analyzed. ***, P < 0.001; **, P < 0.05.
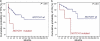
Figure 4.
Kaplan-Meier estimates of treatment-free survival (TFS) and overall survival (OS), according to the mutational status of NOTCH1.
Figure 5.
Timing of NOTCH1 mutations in RS. (a) NOTCH1 mutations in some cases of RS are not identifiable in the preceding CLL phase, whereas in other patients may already be present before transformation, at a clonal/subclonal level. In the heatmap, rows correspond to NOTCH1 mutations and columns represent sequential phases of the disease (paired CLL diagnostic samples and RS samples), color-coded based on the mutational status (for details, see Materials and methods). (b) The NOTCH1 mutation detected in RS case RS63 is present at subclonal levels at the time of CLL diagnosis (sample CLL63). The graph shows a quantitative estimate of the balance between wild-type and mutated (ΔCT7544-7545) NOTCH1 alleles assessed by ultradeep 454 sequencing in the paired CLL-RS samples (CLL63 and RS63). 30 mo elapsed between collection of the CLL sample (diagnosis of CLL) and collection of the RS sample (obtained at diagnosis of RS). Numbers on top indicate the actual number of sequencing reads carrying the mutations over total analyzed (for details, see Materials and methods). (c) Mutations of NOTCH1 are largely mutually exclusive with MYC activating events (translocations and copy number gains) in RS. In the heatmap, rows correspond to the indicated genes, and columns represent individual RS samples, color-coded based on the gene status (white, wild-type; red, mutated or activated or disrupted).
References
- Albesiano E., Messmer B.T., Damle R.N., Allen S.L., Rai K.R., Chiorazzi N. 2003. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 102:3333–3339 10.1182/blood-2003-05-1585 - DOI - PubMed
- Annunziata C.M., Davis R.E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., et al. 2007. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 12:115–130 10.1016/j.ccr.2007.07.004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- R01 CA037295/CA/NCI NIH HHS/United States
- R37 CA037295/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- U54 CA121852-05/CA/NCI NIH HHS/United States
- 1R01LM010140-01/LM/NLM NIH HHS/United States
- R01 LM010140/LM/NLM NIH HHS/United States
- CA-37295/CA/NCI NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases