Distinct functional effects for dynamin 3 during megakaryocytopoiesis - PubMed (original) (raw)
Distinct functional effects for dynamin 3 during megakaryocytopoiesis
Wenjing Wang et al. Stem Cells Dev. 2011 Dec.
Abstract
Dynamin 3 (DNM3) is a member of a family of motor proteins that participate in a number of membrane rearrangements such as cytokinesis, budding of transport vesicles, phagocytosis, and cell motility. Recently, DNM3 was implicated as having a role in megakaryocyte (MK) development. To further investigate the functional role of DNM3 during megakaryocytopoiesis, we introduced sequence-specific short hairpin RNAs (shRNAs) into developing MKs. The results showed that knockdown of DNM3 inhibited a stage of MK development that involved progenitor amplification. This was evident by significant decreases in the number of colony forming unit-megakaryocytes, the total number of nucleated cells, and the number of CD41(+) and CD61(+) MKs produced in culture. Using a styrl membrane dye to quantify the demarcation membrane system (DMS) of terminally differentiated MKs, we found that DNM3 co-localized with the DMS and that DNM3 lentiviral shRNAs precluded the formation of the DMS. Knockdown of dynamin 3 in murine MKs also caused a decrease in the number of morphologically large MKs and the overall size of large MKs was decreased relative to controls. MK protein lysates were used in overlay blots to show that both DNM3 and actin bind to nonmuscle myosin IIA (MYH9). Consistent with these observations, immunofluorescence studies of MKs and proplatelet processes showed co-localization of DNM3 with MYH9. Overall, these studies demonstrate that DNM3 not only participates in MK progenitor amplification, but is also involved in cytoplasmic enlargement and the formation of the DMS.
Figures
FIG. 1.
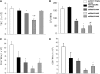
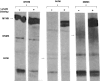
In vitro knockdown validation of _DNM3_-specific shRNAs. (A) Human _DNM3_-specific lentiviral shRNA plasmid sequences. Schematic that represents the coding sequence for human DNM3 and the shRNA targeting sites. (B) RT-PCR was performed to determine the down-regulation levels of DNM3 after UT-7/TPO cells were transfected with the indicated plasmid constructs. (C) Quantitative RT-PCR was performed to confirm the knockdown of DNM3 in culture-derived MKs produced from human UCB CD34+ cells that had been transduced with the indicated shRNA lentiviral vectors. Expression levels are expressed as ratios relative to the shRNA control, which is set to 1. (D) Western blot showed that all 3 lentiviral shRNA constructs knocked down DNM3 protein expression levels. All experiments were performed using cells infected with the pLKO.1 empty vector as a negative control. Three independent experiments were performed. DNM3, dynamin 3; MK, megakaryocyte; shRNA, short hairpin RNA; TPO, thrombopoietin; UCB, umbilical cord blood; PCR, polymerase chain reaction.
FIG. 2.
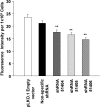
DNM3 knockdown inhibits the proliferation of MK progenitors. Human UCB CD34+ cells were cultured with IL-3, IL-6, stem cell factor, and TPO and transduced with _DNM3_-specific shRNA lentiviral vectors at 24, 48, and 72 h of culture. (A) Total nucleated cell counts after cells were maintained in liquid culture for 10 days. (B) On day 4 of liquid culture, cells were transferred to MegaCult C collagen-based medium and maintained in culture for an additional 10 days. Colonies were stained mouse anti-human GPIIb/IIIa antibody and the total number of colony forming unit-megakaryocyte colonies was determined. (C and D) Total number of CD41+ and CD61+ MKs that is present at day 10 of liquid culture. Data represent the mean±SEM of 4 experiments. Each experiment was conducted in duplicate. *P<0.05, compared to pLKO.1 Empty vector control; **P<0.01, compared to pLKO.1 Empty vector control; #P<0.05, compared to nonspecific shRNA control. IL, interleukin; SEM, standard error of the mean.
FIG. 3.
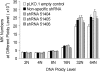
DNA ploidy analysis. UT-7/TPO cells treated with SU6656 were transfected with the indicated plasmids as described in the Materials and Methods section. Ploidy level of UT-7/TPO cells treated with SU6656 and transduced with control or DNM3 shRNA vectors. Data represent the mean±SEM of 4 experiments performed in duplicate. No significant changes in DNA ploidy levels were observed with the knockdown of DNM3.
FIG. 4.
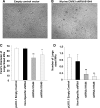
Suppression of dynamin 3 reduces the number of large MKs. Murine BM cells were treated with lentiviral vectors that drive expression of shRNA sequences targeted to murine dynamin 3. On day 4, puromycin (5 μg/mL) was added to cultures to select for transduced cells and on day 7 the cells were observed microscopically. (A) Cells treated with an empty vector control. (B) Cells treated with lentiviral construct driving the expression of dynamin 3-specific shRNA. (C) Overall size of cells as determined by using the “Analyze Particles” command in ImageJ to measure the Feret's diameter. (D) Total number of large cells as determined by flow cytometry. Data represent the mean±SEM of 3 experiments. Each experiment was conducted in duplicate. *P<0.05, compared to the controls; **P<0.01, compared to the controls.
FIG. 5.
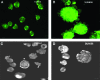
UT-7/TPO cells when treated with SU6656 show a well-developed and invaginated plasma membrane system. Representative confocal and three-dimensional images of UT-7/TPO cells that were treated with and without SU6656 and stained with the styrl membrane dye, di-8-ANEPPS. (A and C) Cells not treated with SU6656. (B and D) Cells treated with SU6656. Scale bar represents 10 μm.
FIG. 6.
Functional knockdown of DNM3 precludes the formation of the MK demarcation membrane system. Relative fluorescence intensity of MKs after staining with di-8-ANEPPS. Data represent the mean±SEM of 2 experiments (_n_=6). **P<0.01 when compared to the controls.
FIG. 7.
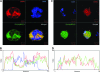
DNM3 is closely associated with the distribution of the DMS in MKs. (A) UT-7/TPO cells treated with SU6656 and transfected with a DNM3_mCherry lentiviral fusion vector were stained with DAPI, and the styrl membrane dye, di-8-ANEPPS. (B) UCB CD34+ cells were cultured in MK induction media as described in the Materials and Methods section. On day 7, CD41+ cell were isolated by flow cytometry and the CD41+ cells were cultured for an additional 3 days prior to staining with DAPI and di-8-ANEPPS. (C and D) Graphic depiction of cross-sectional line on merged images. Red, DNM3; green, DMS stained by di-8-AMEPPS; and blue, DAPI. Three independent experiments were performed. DMS, demarcation membrane system.
FIG. 8.
DNM3 and actin bind to immunoprecipitated MYH9 in UT-7/TPO cells. UT-7/TPO protein lysates were immunoprecipitated with anti-myosin IIA antibody. Eluted fractions were first separated on sodium dodecyl sulfate– polyacrylamide gel electrophoresis followed by transfer of the protein onto polyvinylidene fluoride membrane. After blocking, the protein lanes were cut into strips. For each antibody used in the experiment one lane remained in blocking solution (control), and a second strip for each antibody was incubated in fresh UT-7/TPO lysate (O/L=overlay). All strips were washed with TBS+0.05% Triton and immunostained with indicated antibodies.
FIG. 9.
Co-localization of DNM3 and MYH9. (A) UT-7/TPO cells treated with SU6656. (B) Histogram of cross-sectional line depicted on merged image of UT-7/TPO cells. (C) UCB CD34+ cells cultured in MK induction media for 7 days, sorted for CD41+ cells, and cultured for an additional 3 days. (D) Histogram of cross-sectional line depicted on merged image of primary cultured-derived MKs. (E) Proplatelet processes extending from primary human cultured-derived MKs. (F) Histogram of cross-sectional line depicted on merged image of proplatelet processes. Red, DNM3; green, MYH9.
Similar articles
- Dynamins 2 and 3 control the migration of human megakaryocytes by regulating CXCR4 surface expression and ITGB1 activity.
Suraneni PK, Corey SJ, Hession MJ, Ishaq R, Awomolo A, Hasan S, Shah C, Liu H, Wickrema A, Debili N, Crispino JD, Eklund EA, Chen Y. Suraneni PK, et al. Blood Adv. 2018 Dec 11;2(23):3540-3552. doi: 10.1182/bloodadvances.2018021923. Blood Adv. 2018. PMID: 30538113 Free PMC article. - Dynamin 3 participates in the growth and development of megakaryocytes.
Reems JA, Wang W, Tsubata K, Abdurrahman N, Sundell B, Tijssen MR, van der Schoot E, Di Summa F, Patel-Hett S, Italiano J Jr, Gilligan DM. Reems JA, et al. Exp Hematol. 2008 Dec;36(12):1714-27. doi: 10.1016/j.exphem.2008.08.010. Exp Hematol. 2008. PMID: 19007685 Free PMC article. - Myosin IIA is critical for organelle distribution and F-actin organization in megakaryocytes and platelets.
Pertuy F, Eckly A, Weber J, Proamer F, Rinckel JY, Lanza F, Gachet C, Léon C. Pertuy F, et al. Blood. 2014 Feb 20;123(8):1261-9. doi: 10.1182/blood-2013-06-508168. Epub 2013 Nov 15. Blood. 2014. PMID: 24243973 - MicroRNA function in megakaryocytes.
Raghuwanshi S, Dahariya S, Musvi SS, Gutti U, Kandi R, Undi RB, Sahu I, Gautam DK, Paddibhatla I, Gutti RK. Raghuwanshi S, et al. Platelets. 2019;30(7):809-816. doi: 10.1080/09537104.2018.1528343. Epub 2018 Oct 25. Platelets. 2019. PMID: 30359163 Review. - Hierarchical structure of human megakaryocyte progenitor cells.
Hoffman R, Murrav LJ, Young JC, Luens KM, Bruno E. Hoffman R, et al. Stem Cells. 1996;14 Suppl 1:75-81. doi: 10.1002/stem.5530140709. Stem Cells. 1996. PMID: 11012205 Review.
Cited by
- Dynamin-related protein-1 controls fusion pore dynamics during platelet granule exocytosis.
Koseoglu S, Dilks JR, Peters CG, Fitch-Tewfik JL, Fadel NA, Jasuja R, Italiano JE Jr, Haynes CL, Flaumenhaft R. Koseoglu S, et al. Arterioscler Thromb Vasc Biol. 2013 Mar;33(3):481-8. doi: 10.1161/ATVBAHA.112.255737. Epub 2013 Jan 3. Arterioscler Thromb Vasc Biol. 2013. PMID: 23288151 Free PMC article. - Platelets and cancer: a casual or causal relationship: revisited.
Menter DG, Tucker SC, Kopetz S, Sood AK, Crissman JD, Honn KV. Menter DG, et al. Cancer Metastasis Rev. 2014 Mar;33(1):231-69. doi: 10.1007/s10555-014-9498-0. Cancer Metastasis Rev. 2014. PMID: 24696047 Free PMC article. Review. - New insights of platelet endocytosis and its implication for platelet function.
Zhou Y, Dong J, Wang M, Liu Y. Zhou Y, et al. Front Cardiovasc Med. 2024 Jan 9;10:1308170. doi: 10.3389/fcvm.2023.1308170. eCollection 2023. Front Cardiovasc Med. 2024. PMID: 38264257 Free PMC article. Review. - Dynamins 2 and 3 control the migration of human megakaryocytes by regulating CXCR4 surface expression and ITGB1 activity.
Suraneni PK, Corey SJ, Hession MJ, Ishaq R, Awomolo A, Hasan S, Shah C, Liu H, Wickrema A, Debili N, Crispino JD, Eklund EA, Chen Y. Suraneni PK, et al. Blood Adv. 2018 Dec 11;2(23):3540-3552. doi: 10.1182/bloodadvances.2018021923. Blood Adv. 2018. PMID: 30538113 Free PMC article. - Enhancing functional platelet release in vivo from in vitro-grown megakaryocytes using small molecule inhibitors.
Jarocha D, Vo KK, Lyde RB, Hayes V, Camire RM, Poncz M. Jarocha D, et al. Blood Adv. 2018 Mar 27;2(6):597-606. doi: 10.1182/bloodadvances.2017010975. Blood Adv. 2018. PMID: 29545255 Free PMC article.
References
- Orth JD. McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–39. - PubMed
- Okamoto PM. Herskovits JS. Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272:11629–11635. - PubMed
- Krutchen AE. McNiven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119(Pt 9):1683–1690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous