Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation - PubMed (original) (raw)
Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation
Stuart A Sievers et al. Nature. 2011.
Abstract
Many globular and natively disordered proteins can convert into amyloid fibrils. These fibrils are associated with numerous pathologies as well as with normal cellular functions, and frequently form during protein denaturation. Inhibitors of pathological amyloid fibril formation could be useful in the development of therapeutics, provided that the inhibitors were specific enough to avoid interfering with normal processes. Here we show that computer-aided, structure-based design can yield highly specific peptide inhibitors of amyloid formation. Using known atomic structures of segments of amyloid fibrils as templates, we have designed and characterized an all-D-amino-acid inhibitor of the fibril formation of the tau protein associated with Alzheimer's disease, and a non-natural L-amino-acid inhibitor of an amyloid fibril that enhances sexual transmission of human immunodeficiency virus. Our results indicate that peptides from structure-based designs can disrupt the fibril formation of full-length proteins, including those, such as tau protein, that lack fully ordered native structures. Because the inhibiting peptides have been designed on structures of dual-β-sheet 'steric zippers', the successful inhibition of amyloid fibril formation strengthens the hypothesis that amyloid spines contain steric zippers.
©2011 Macmillan Publishers Limited. All rights reserved
Conflict of interest statement
The authors declare no competing financial interests.
Figures
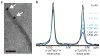
Figure 1. Scheme for the design and characterization of peptide inhibitors of amyloid fibrillation
Tau constructs form fibers in vitro (top left). The VQIVYK segment in isolation forms fibers and microcrystals (bottom left). The atomic structure of the fiber-like VQIVYK segment reveals a characteristic steric zipper motif, comprising a pair of interacting β-sheets running along the fiber axis (grey arrow), in purple and grey (bottom right). We designed a D-amino acid peptide to bind to the end of the steric zipper template and prevent fiber elongation (middle right). The D-peptide, in red, is designed to satisfy hydrogen bonds and make favorable apolar interactions with the molecule below, while preventing the addition of other molecules above and on the opposite β-sheet. As shown in vitro, the designed D-peptide prevents the formation of fibers when incubated with tau K19 (upper right). Scale bars are 100 μm and 200 nm on the microcrystal image and electron micrographs, respectively.
Figure 2. Designed D-peptide delays tau K12 fibrillation in a sequence-specific manner
a, Tau construct composition. The largest human tau isoform hTau40 contains four microtubule-binding repeats, R1 – R4, while K12 and K19 lack R2. The black bars at the N-termini of R2 and R3 represent the fibrillogenic segments VQIINK and VQIVYK, respectively. b, The inhibitor D-TLKIVW is designed to interact with atoms on both β-strands of the VQIVYK steric zipper primarily through hydrophobic packing and hydrogen bonding interactions. c, The inhibitor (red) interacts with the VQIVYK structure below (grey). The transparent spheres show where the two molecules interact favorably. Black and red dashes indicate main chain and side chain hydrogen bonds, respectively. d, The seeded fibrillation of 50 μM K12 in the presence and absence of ten-fold molar excess peptide was monitored by Thioflavin S fluorescence. In the presence of the scrambled peptide D-TIWKVL (dark green) or alone (black), seeded K12 fibrillation occurs with almost no lag time. However, D-TLKIVW prevents fibrillation for days (maroon). e, At equimolar concentrations, D-TLKIVW (red) inhibits the fibrillation of 50μM K12. D-TIKWVL (blue) with only three residues scrambled shows weak inhibition. However, no inhibition is observed for either D-TIWKVL (green) or D-LKTWIV (cyan). f, The replacement of D-Leu2, designed to clash with VQIVYK on the opposite sheet, with D-Ala eliminates the inhibition of fibrillation.
Figure 3. Mechanism of interaction
a, Nanogold covalently bound to D-TLKIVW localizes at the ends of two tau K19 fibers. Scale bar represents 50 nm. b, The inhibitor D-TLKIVW binds to fibers with an estimated affinity constant in the low micromolar range, as shown by the indole proton region of the 500 MHz 1H NMR spectra of D-TLKIVW (9.83 ppm) and D-LKTWIV (9.98 ppm) in the presence of increasing concentrations of VQIVYK fibers. The resonance of the D-TLKIVW indole proton is reduced in the presence of increasing concentrations of VQIVYK fibers, whereas the indole proton signal for the scrambled control peptide D-LKTWIV is only slightly affected. Fiber solutions contain 0 to 1500 μM of VQIVYK monomers, as indicated.
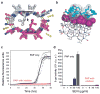
Figure 4. A designed non-natural peptide inhibits 248PAP286 fibrillation
a, The view down the fiber axis of the crystal structure of the GGVLVN steric zipper reveals two mating β-sheets with parallel, in-register β-strands (hydrogen bonds depicted as green dashed lines; water molecules as yellow spheres). b, View roughly perpendicular to a fiber of 3 layers, with the atoms of the side-chains of the top layer shown as purple spheres. On top, in aqua, is a designed non-natural peptide inhibitor, W-H-K-chAla-W-hydroxyTic (see Supplementary Fig. 13). c, The inhibitor blocks 248PAP286 fibrillation, as shown by monitoring Thioflavin T fluorescence. With two-fold molar excess of the inhibitor (red), the fluorescence remains low over the course of the experiment for all five replicates, in contrast to 248PAP286 alone (black). d, HIV infection rates were determined by monitoring β-galactosidase activity. Agitated 248PAP286 (SEVI) alone efficiently increases viral infection, whereas 248PAP286 mixtures incubated with inhibitor were unable to enhance HIV infection. Peptide concentrations during virion treatment are indicated on the x-axis. Error bars show the s.d. of 3 measurements per sample.
Similar articles
- Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.
Nguyen P, Derreumaux P. Nguyen P, et al. Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review. - Structure and intermolecular dynamics of aggregates populated during amyloid fibril formation studied by hydrogen/deuterium exchange.
Carulla N, Zhou M, Giralt E, Robinson CV, Dobson CM. Carulla N, et al. Acc Chem Res. 2010 Aug 17;43(8):1072-9. doi: 10.1021/ar9002784. Acc Chem Res. 2010. PMID: 20557067 - Polymorphic fibril formation by residues 10-40 of the Alzheimer's beta-amyloid peptide.
Paravastu AK, Petkova AT, Tycko R. Paravastu AK, et al. Biophys J. 2006 Jun 15;90(12):4618-29. doi: 10.1529/biophysj.105.076927. Epub 2006 Mar 24. Biophys J. 2006. PMID: 16565054 Free PMC article. - Recent Advances by In Silico and In Vitro Studies of Amyloid-β 1-42 Fibril Depicted a S-Shape Conformation.
Villalobos Acosta DMÁ, Chimal Vega B, Correa Basurto J, Fragoso Morales LG, Rosales Hernández MC. Villalobos Acosta DMÁ, et al. Int J Mol Sci. 2018 Aug 16;19(8):2415. doi: 10.3390/ijms19082415. Int J Mol Sci. 2018. PMID: 30115846 Free PMC article. Review.
Cited by
- NMR Meets Tau: Insights into Its Function and Pathology.
Lippens G, Landrieu I, Smet C, Huvent I, Gandhi NS, Gigant B, Despres C, Qi H, Lopez J. Lippens G, et al. Biomolecules. 2016 Jun 7;6(2):28. doi: 10.3390/biom6020028. Biomolecules. 2016. PMID: 27338491 Free PMC article. Review. - Fibril-forming motifs are essential and sufficient for the fibrillization of human Tau.
Meng SR, Zhu YZ, Guo T, Liu XL, Chen J, Liang Y. Meng SR, et al. PLoS One. 2012;7(6):e38903. doi: 10.1371/journal.pone.0038903. Epub 2012 Jun 11. PLoS One. 2012. PMID: 22701727 Free PMC article. - Synthetic phosphoethanolamine-modified oligosaccharides reveal the importance of glycan length and substitution in biofilm-inspired assemblies.
Tyrikos-Ergas T, Gim S, Huang JY, Pinzón Martín S, Varón Silva D, Seeberger PH, Delbianco M. Tyrikos-Ergas T, et al. Nat Commun. 2022 Jul 8;13(1):3954. doi: 10.1038/s41467-022-31633-5. Nat Commun. 2022. PMID: 35804023 Free PMC article. - The Rational Discovery of a Tau Aggregation Inhibitor.
Baggett DW, Nath A. Baggett DW, et al. Biochemistry. 2018 Oct 23;57(42):6099-6107. doi: 10.1021/acs.biochem.8b00581. Epub 2018 Oct 5. Biochemistry. 2018. PMID: 30247897 Free PMC article. - Self-Assembly of Aromatic Amino Acid Enantiomers into Supramolecular Materials of High Rigidity.
Bera S, Xue B, Rehak P, Jacoby G, Ji W, Shimon LJW, Beck R, Král P, Cao Y, Gazit E. Bera S, et al. ACS Nano. 2020 Feb 25;14(2):1694-1706. doi: 10.1021/acsnano.9b07307. Epub 2020 Jan 21. ACS Nano. 2020. PMID: 31944667 Free PMC article.
References
- Westermark P, et al. A primer of amyloid nomenclature. Amyloid. 2007;14:179–183. - PubMed
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources