Genetics and pathogenesis of inflammatory bowel disease - PubMed (original) (raw)
Review
Genetics and pathogenesis of inflammatory bowel disease
Bernard Khor et al. Nature. 2011.
Abstract
Recent advances have provided substantial insight into the maintenance of mucosal immunity and the pathogenesis of inflammatory bowel disease. Cellular programs responsible for intestinal homeostasis use diverse intracellular and intercellular networks to promote immune tolerance, inflammation or epithelial restitution. Complex interfaces integrate local host and microbial signals to activate appropriate effector programs selectively and even drive plasticity between these programs. In addition, genetic studies and mouse models have emphasized the role of genetic predispositions and how they affect interactions with microbial and environmental factors, leading to pro-colitogenic perturbations of the host-commensal relationship.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
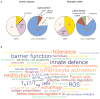
Figure 1. Genetic architecture of IBD-linked susceptibility loci
a, GWAS have identified 71 risk loci in Crohn’s disease and 47 risk loci in ulcerative colitis (P value of association < 5×10−8). Of these, 28 risk loci exhibit shared associations (defined as _P_ < 5×10−8 for either Crohn’s disease or ulcerative colitis, and _P_ < 1×10−4 for the other form of IBD). Approximately half of the loci implicated in Crohn’s disease and ulcerative colitis are associated with _cis_- and/or _trans_-expression quantitative trait loci (eQTL) effects (left panels). Genes whose expression are affected by these variants could also be involved in IBD pathogenesis. The loci composition (right panels) shows the number of genes that either lie within or segregate in linkage disequilibrium with IBD-implicated loci (coefficient of correlation _r_2 > 0.8). These loci are structurally heterogeneous, and are associated with widely ranging numbers of genes. Loci not associated with any genes, known as gene deserts, frequently contain non-coding transcripts or predicted open reading frames (ORFs), and can be associated with _trans_-eQTL effects. b, Recurring terms illustrating biological processes implicated by at least three genes represented in IBD loci; font sizes are proportional to the number of genes associated with each respective process. Breg cells, B regulatory cells; ER, endoplasmic reticulum; GPCR, G-protein-coupled receptor; IL, interleukin; lincRNA, large intervening non-coding RNA; miRNA, microRNA; ncRNA, non-coding RNA; NF-κB, nuclear factor-κB; ROS, reactive oxygen species; TH17 cells, T helper 17 cells; Treg cells, T regulatory cells.
Figure 2. A model for IBD pathways based on GWAS
Intestinal homeostasis involves the coordinated actions of epithelial, innate and adaptive immune cells. Barrier permeability permits microbial incursion, which is detected by the innate immune system, which then orchestrates appropriate tolerogenic, inflammatory and restitutive responses in part by releasing extracellular mediators that recruit other cellular components, including adaptive immune cells. Genetic variants, the microbiota and immune factors affect the balance of these signals. Genes in linkage disequilibrium (_r_2 > 0.8) with IBD-associated single nucleotide polymorphisms (SNPs) were manually curated and classified according to their function(s) in the context of intestinal homeostasis and immunity. Text colour indicates whether the genes are linked to risk loci associated with Crohn’s disease (CD; black), ulcerative colitis (UC; blue) or both (red). Asterisk denotes corresponding coding mutations; _cis_-eQTL effects are underlined. G, goblet cell; P, Paneth cell.
Figure 3. Genetic variants in IBD signalling modules
Schematic of selected signalling pathways involved in the maintenance of intestinal homeostasis, including epithelial junctional complex assembly, innate immune recognition of pathogen-associated motifs, GPCRs and immune defence, anti-inflammatory interleukin-10 (IL-10) signalling, TH17-cell differentiation, inhibitory pathways in lymphocyte signalling, and B-cell activation and IgA antibody responses. Proteins encoded by genes identified as being in linkage disequilibrium with IBD-risk SNPs (_r_2 > 0.8) are highlighted in red. BCR, B-cell receptor; cAMP, cyclic AMP; EGFR, epidermal growth factor receptor; ERRFI1, ERBB receptor feedback inhibitor 1; Gα12, G protein subunit α12; GPCR, G-protein-coupled receptor; HNF-4α, hepatocyte nuclear factor-4α; LPA, lysophosphatidic acid; MDP, muramyl dipeptide; NF-κB, nuclear factor-κB; PI(3)K, phosphatidylinositol-3-OH kinase; PLA2G2E, phospholipase A2, group IIE; PTGER4, prostaglandin E receptor 4; SCFA, short-chain fatty acid; SIAE, sialic acid acetylesterase; ssRNA, single-stranded RNA; TCR, T-cell receptor.
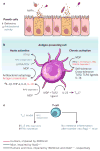
Figure 4. Cell-intrinsic functions of NOD2
NOD2 is activated by the bacterial peptidoglycan muramyl dipeptide (MDP). Cell-specific NOD2 functions are shown, distinguishing between those functions impaired in cells from humans with the Crohn’s-disease-associated mutation 3020insC (red), from _Nod2_-deficient mice (blue), or from both (black). a, In Paneth cells, Nod2 deficiency leads to attenuated antibacterial activity in the intestinal crypts and decreased expression of α-defensin 4 (encoded by Defcr4, also known as Defa4) and α-defensin-related sequence 10 (DEFCR-RS10, also known as DEFA-RS10). b, MDP-stimulated release of pro-inflammatory NF-κB-dependent cytokines (such as IL-1β, TNF-α and IL-6), as well as secretion of IL-23 (which promotes TH17 differentiation) after co-stimulation with MDP and TLR2 ligands, is decreased in antigen-presenting cells from _Nod2_-deficient mice or 3020insC human donors. MDP stimulation also leads to NOD2-activated autophagy and antigen presentation. In mice, the activation of antigen-presenting cells by ssRNA or respiratory syncytial virus (RSV) stimulates secretion of type I interferon (IFN-β) in a NOD2-dependent, receptor-interacting protein-2 (RIP2)-independent fashion. In contrast to the pro-inflammatory effects, chronic NOD2 activation (right) by MDP induces both self-tolerance and cross-tolerance to IL-1β, and TLR2 and TLR4 ligands. This is dependent on IRF4 in mice and humans, and also on IL-10, TGF-β, IL-1RA and IL-1R-associated kinase M (IRAK-M) in humans. MDP-induced tolerance is lost in _Nod2_-deficient mice and in patients with the 3020insC variant. NOD2-dependent release of IL-10 after MDP stimulation has been demonstrated to be specific to humans and is impaired in 3020insC cells. MDP-stimulated release of several cytokines, including IL-10, IL-1β, TNF-α and IL-6, is dependent on RIP2. c, In mice, NOD2 mediates IFN-γ secretion and REL-dependent IL-2 production in T cells in response to Toxoplasma gondii infection. Also, Nod2 deficiency attenuates the ability of T cells to cause experimental colitis after transfer into _Rag1_-deficient hosts.
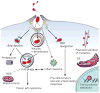
Figure 5. Intracellular defence programs in microbial recognition
Host cells have evolved processes by which they restrict the availability of intracellular permissive niches to microbes. Microbial recognition by PRRs, such as NOD proteins and TLRs, activates key immediate host programs, leading to polarized secretion of pro-inflammatory mediators (directed to either the luminal or basolateral surface). Bacteria can either be maintained in subcellular compartments such as microbe-containing vacuoles, or escape into the cytoplasm, where they can be ubiquitylated and targeted for degradation. Both subsets can be targeted by the autophagy pathway, which is also regulated by other host defence mechanisms such as oxidative stress and inflammasome activation.
Similar articles
- Unravelling the pathogenesis of inflammatory bowel disease.
Xavier RJ, Podolsky DK. Xavier RJ, et al. Nature. 2007 Jul 26;448(7152):427-34. doi: 10.1038/nature06005. Nature. 2007. PMID: 17653185 Review. - Immunopathophysiology of inflammatory bowel disease: how genetics link barrier dysfunction and innate immunity to inflammation.
Mehta M, Ahmed S, Dryden G. Mehta M, et al. Innate Immun. 2017 Aug;23(6):497-505. doi: 10.1177/1753425917722206. Innate Immun. 2017. PMID: 28770665 Review. - Intestinal microbiota in inflammatory bowel disease: friend of foe?
Fava F, Danese S. Fava F, et al. World J Gastroenterol. 2011 Feb 7;17(5):557-66. doi: 10.3748/wjg.v17.i5.557. World J Gastroenterol. 2011. PMID: 21350704 Free PMC article. - Intestinal homeostasis and its breakdown in inflammatory bowel disease.
Maloy KJ, Powrie F. Maloy KJ, et al. Nature. 2011 Jun 15;474(7351):298-306. doi: 10.1038/nature10208. Nature. 2011. PMID: 21677746 Review.
Cited by
- Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea.
Guard BC, Barr JW, Reddivari L, Klemashevich C, Jayaraman A, Steiner JM, Vanamala J, Suchodolski JS. Guard BC, et al. PLoS One. 2015 May 22;10(5):e0127259. doi: 10.1371/journal.pone.0127259. eCollection 2015. PLoS One. 2015. PMID: 26000959 Free PMC article. - Comparative Study of Intestinal Microbiome in Patients with Ulcerative Colitis and Healthy Controls in Korea.
Do KH, Ko SH, Kim KB, Seo K, Lee WK. Do KH, et al. Microorganisms. 2023 Nov 11;11(11):2750. doi: 10.3390/microorganisms11112750. Microorganisms. 2023. PMID: 38004761 Free PMC article. - The immunological landscape in necrotising enterocolitis.
Cho SX, Berger PJ, Nold-Petry CA, Nold MF. Cho SX, et al. Expert Rev Mol Med. 2016 Jun 24;18:e12. doi: 10.1017/erm.2016.13. Expert Rev Mol Med. 2016. PMID: 27341512 Free PMC article. Review. - CX(3)CR1(+) macrophages support IL-22 production by innate lymphoid cells during infection with Citrobacter rodentium.
Manta C, Heupel E, Radulovic K, Rossini V, Garbi N, Riedel CU, Niess JH. Manta C, et al. Mucosal Immunol. 2013 Jan;6(1):177-88. doi: 10.1038/mi.2012.61. Epub 2012 Aug 1. Mucosal Immunol. 2013. PMID: 22854708 Free PMC article. - Use of Humanized Mice to Study the Pathogenesis of Autoimmune and Inflammatory Diseases.
Koboziev I, Jones-Hall Y, Valentine JF, Webb CR, Furr KL, Grisham MB. Koboziev I, et al. Inflamm Bowel Dis. 2015 Jul;21(7):1652-73. doi: 10.1097/MIB.0000000000000446. Inflamm Bowel Dis. 2015. PMID: 26035036 Free PMC article. Review.
References
- Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature Genet. 2010;42:1118–1125. This study identifies 39 new loci containing genes in previously identified Crohn’s-disease-relevant pathways, including IL10, IL27 and TYK2, and reports the important genetic overlap with loci implicated in other immune-related diseases. - PMC - PubMed
- Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature Genet. 2011;43:246–252. This study identifies 29 new loci containing genes that reinforce the contribution of epithelial barrier function, intracellular defence and cytokine-dependent signalling in ulcerative colitis, and it also reports genetic overlap with Crohn’s disease. - PMC - PubMed
- Spehlmann ME, et al. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–976. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- R01 DK083756-04/DK/NIDDK NIH HHS/United States
- P30 DK040561-15/DK/NIDDK NIH HHS/United States
- T32 CA009216/CA/NCI NIH HHS/United States
- R01 DK083756/DK/NIDDK NIH HHS/United States
- P30 DK043351-20/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources