Ammonia-induced autophagy is independent of ULK1/ULK2 kinases - PubMed (original) (raw)
Ammonia-induced autophagy is independent of ULK1/ULK2 kinases
Heesun Cheong et al. Proc Natl Acad Sci U S A. 2011.
Erratum in
- Proc Natl Acad Sci U S A. 2011 Oct 25;108(43):17856
Abstract
Autophagy, a lysosome-mediated catabolic process, contributes to maintenance of intracellular homeostasis and cellular response to metabolic stress. In yeast, genes essential to the execution of autophagy have been defined, including autophagy-related gene 1 (ATG1), a kinase responsible for initiation of autophagy downstream of target of rapamycin. Here we investigate the role of the mammalian Atg1 homologs, uncoordinated family member (unc)-51-like kinase 1 and 2 (ULK1 and ULK2), in autophagy by generating mouse embryo fibroblasts (MEFs) doubly deficient for ULK1 and ULK2. We found that ULK1/2 are required in the autophagy response to amino acid deprivation but not for autophagy induced by deprivation of glucose or inhibition of glucose metabolism. This ULK1/2-independent autophagy was not the simple result of bioenergetic compromise and failed to be induced by AMP-activated protein kinase activators such as 5-aminoimidazole-4-carboxamide riboside and phenformin. Instead we found that autophagy induction upon glucose deprivation correlated with a rise in cellular ammonia levels caused by elevated amino acid catabolism. Even in complete medium, ammonia induced autophagy in WT and Ulk1/2(-/-) MEFs but not in Atg5-deficient MEFs. The autophagy response to ammonia is abrogated by a cell-permeable form of pyruvate resulting from the scavenging of excess ammonia through pyruvate conversion to alanine. Thus, although ULK1 and/or ULK2 are required for the autophagy response following deprivation of nitrogenous amino acids, the autophagy response to the enhanced amino acid catabolism induced by deprivation of glucose or direct exposure to ammonia does not require ULK1 and/or ULK2. Together, these data suggest that autophagy provides cells with a mechanism to adapt not only to nitrogen deprivation but also to nitrogen excess.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
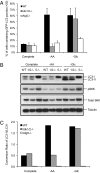
Ulk1/2 are required for the autophagy response to amino acid deprivation but not for the response to glucose deprivation. GFP-LC3–expressing WT, Ulk1/2 DKO (Ulk1/2−/−) and Atg5 KO (Atg5−/−) MEFs were cultured in complete medium for 24 h and then were left untreated or were subjected to amino acid starvation (−AA) for 6 h or glucose starvation (−Glc) for 24 h as described in Materials and Methods. (A) Autophagy activity was measured by fluorescence microscopy. The percentage of cells containing >20 GFP-LC3 puncta was determined. (B) LC3 conversion from cytosolic LC3 (LC3-I) to lipid-conjugated LC3 (LC3-II) and the presence of phopho-p70 S6 kinase (pS6K), total p70 S6 kinase (S6K), and tubulin were detected by immunoblotting. (C) The conversion ratio of LC3-II/LC3-I of the experiment shown in B was quantified by Image J (National Institutes of Health).
Fig. 2.
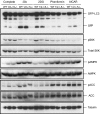
Ulk1/2 are not required for autophagy in response to bioenergetic stress. WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium and then were subjected to glucose starvation (−Glc) or treatment with 10 mM 2DG, 1 mM phenformin, or 2 mM AICAR for 24 h. GFP-LC3 processing was determined by immunoblotting to monitor autophagy. Immunoblotting for phopho-p70 S6 kinase (pS6K), p70 S6 kinase (S6K), phospho-AMPK (pAMPK), total AMPK, phospho-ACC (pACC), total ACC, and tubulin was performed also. Asterisk indicates nonspecific bands.
Fig. 3.
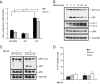
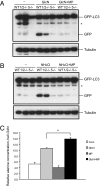
Ammonia stimulates mTOR-independent autophagy. (A) Glucose deprivation, but not amino acid starvation, increases ammonia production. Ammonia levels in media of cells cultured for 24 h were determined with the Nova Biomedical Flex Analyzer. *P < 0.05. (B) NH4Cl treatment increases autophagy without affecting mTOR activity. WT MEFs expressing GFP-LC3 were treated with varying concentrations of NH4Cl in complete medium for 24 h. Autophagy was determined by GFP-LC3 processing and LC3 conversion. mTOR activity also was assessed by immunoblotting for phospho-p70 S6 kinase (pS6K). (C) Atg5, but not Ulk1/2, is required for ammonia-induced autophagy. The GFP-LC3–expressing WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs were treated with 2 mM NH4Cl for 24 h. Autophagy was measured by immunoblotting for GFP-LC3 processing. (D) The dose of NH4Cl that induces autophagy has no effect on cell viability. After 48 h incubation at the conditions indicated in C, cell viability was determined by propidium iodide (PI) exclusion.
Fig. 4.
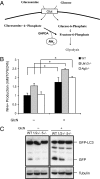
Ammonia generated following glucosamine treatment induces autophagy. (A) Diagram showing ammonia production during glucosamine metabolism. (B) WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium with 10 mM glucosamine (GlcN) for 24 h. Ammonia levels were measured in supernatants of cell cultures with the Nova Biomedical Flex Analyzer. *P < 0.05. (C) Glucosamine treatment increases autophagy. Autophagy was determined by GFP-LC3 processing at 24 h as indicated in B.
Fig. 5.
MP abrogates autophagy induced by ammonia. Autophagy induction by (A) glucosamine (GlcN) or (B) NH4Cl is suppressed by MP treatment. WT, Ulk1/2 DKO (1/2−/−), and Atg5 KO (5−/−) MEFs expressing GFP-LC3 were cultured in complete medium for 24 h before drug treatments. MP (20 mM) was added for an additional 24 h with either 10 mM glucosamine (A) or 2 mM NH4Cl (B). GFP-LC3 processing was detected by immunoblotting to assess autophagy. (C) Relative alanine levels in the culture media of the cells following 24 h incubation as indicated in A were measured by GC-MS analysis in WT MEFs. Com, complete medium. *P < 0.05.
Similar articles
- Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes.
Ro SH, Jung CH, Hahn WS, Xu X, Kim YM, Yun YS, Park JM, Kim KH, Seo M, Ha TY, Arriaga EA, Bernlohr DA, Kim DH. Ro SH, et al. Autophagy. 2013 Dec;9(12):2103-14. doi: 10.4161/auto.26563. Epub 2013 Oct 8. Autophagy. 2013. PMID: 24135897 Free PMC article. - The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy.
Lee EJ, Tournier C. Lee EJ, et al. Autophagy. 2011 Jul;7(7):689-95. doi: 10.4161/auto.7.7.15450. Epub 2011 Jul 1. Autophagy. 2011. PMID: 21460635 Free PMC article. - The autophagy-inducing kinases, ULK1 and ULK2, regulate axon guidance in the developing mouse forebrain via a noncanonical pathway.
Wang B, Iyengar R, Li-Harms X, Joo JH, Wright C, Lavado A, Horner L, Yang M, Guan JL, Frase S, Green DR, Cao X, Kundu M. Wang B, et al. Autophagy. 2018;14(5):796-811. doi: 10.1080/15548627.2017.1386820. Epub 2017 Dec 24. Autophagy. 2018. PMID: 29099309 Free PMC article. - The role of the Atg1/ULK1 complex in autophagy regulation.
Mizushima N. Mizushima N. Curr Opin Cell Biol. 2010 Apr;22(2):132-9. doi: 10.1016/j.ceb.2009.12.004. Epub 2010 Jan 6. Curr Opin Cell Biol. 2010. PMID: 20056399 Review. - Canonical and noncanonical functions of ULK/Atg1.
Wang B, Kundu M. Wang B, et al. Curr Opin Cell Biol. 2017 Apr;45:47-54. doi: 10.1016/j.ceb.2017.02.011. Epub 2017 Mar 11. Curr Opin Cell Biol. 2017. PMID: 28292700 Free PMC article. Review.
Cited by
- Non-autophagic Golgi-LC3 lipidation facilitates TFE3 stress response against Golgi dysfunction.
Kang J, Li CM, Kim N, Baek J, Jung YK. Kang J, et al. EMBO J. 2024 Nov;43(21):5085-5113. doi: 10.1038/s44318-024-00233-y. Epub 2024 Sep 16. EMBO J. 2024. PMID: 39284911 Free PMC article. - Differential and convergent utilization of autophagy components by positive-strand RNA viruses.
Abernathy E, Mateo R, Majzoub K, van Buuren N, Bird SW, Carette JE, Kirkegaard K. Abernathy E, et al. PLoS Biol. 2019 Jan 4;17(1):e2006926. doi: 10.1371/journal.pbio.2006926. eCollection 2019 Jan. PLoS Biol. 2019. PMID: 30608919 Free PMC article. - Fundc1-dependent mitophagy is obligatory to ischemic preconditioning-conferred renoprotection in ischemic AKI via suppression of Drp1-mediated mitochondrial fission.
Wang J, Zhu P, Li R, Ren J, Zhou H. Wang J, et al. Redox Biol. 2020 Feb;30:101415. doi: 10.1016/j.redox.2019.101415. Epub 2019 Dec 28. Redox Biol. 2020. PMID: 31901590 Free PMC article. - Autophagy in Pulmonary Innate Immunity.
Rao L, Eissa NT. Rao L, et al. J Innate Immun. 2020;12(1):21-30. doi: 10.1159/000497414. Epub 2019 Apr 24. J Innate Immun. 2020. PMID: 31018206 Free PMC article. Review. - Conservation of structure, function and inhibitor binding in UNC-51-like kinase 1 and 2 (ULK1/2).
Chaikuad A, Koschade SE, Stolz A, Zivkovic K, Pohl C, Shaid S, Ren H, Lambert LJ, Cosford NDP, Brandts CH, Knapp S. Chaikuad A, et al. Biochem J. 2019 Mar 12;476(5):875-887. doi: 10.1042/BCJ20190038. Biochem J. 2019. PMID: 30782972 Free PMC article.
References
- Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. - PubMed
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials