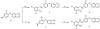Discovery and structure-activity relationships of modified salicylanilides as cell permeable inhibitors of poly(ADP-ribose) glycohydrolase (PARG) - PubMed (original) (raw)
. 2011 Aug 11;54(15):5403-13.
doi: 10.1021/jm200325s. Epub 2011 Jul 8.
Affiliations
- PMID: 21692479
- PMCID: PMC3150619
- DOI: 10.1021/jm200325s
Discovery and structure-activity relationships of modified salicylanilides as cell permeable inhibitors of poly(ADP-ribose) glycohydrolase (PARG)
Jamin D Steffen et al. J Med Chem. 2011.
Abstract
The metabolism of poly(ADP-ribose) (PAR) in response to DNA strand breaks, which involves the concerted activities of poly(ADP-ribose) polymerases (PARPs) and poly(ADP-ribose) glycohydrolase (PARG), modulates cell recovery or cell death depending upon the level of DNA damage. While PARP inhibitors show high promise in clinical trials because of their low toxicity and selectivity for BRCA related cancers, evaluation of the therapeutic potential of PARG is limited by the lack of well-validated cell permeable inhibitors. In this study, target-related affinity profiling (TRAP), an alternative to high-throughput screening, was used to identify a number of druglike compounds from several chemical classes that demonstrated PARG inhibition in the low-micromolar range. A number of analogues of one of the most active chemotypes were synthesized to explore the structure-activity relationship (SAR) for that series. This led to the discovery of a putative pharmacophore for PARG inhibition that contains a modified salicylanilide structure. Interestingly, these compounds also inhibit PARP-1, indicating strong homology in the active sites of PARG and PARP-1 and raising a new challenge for development of PARG specific inhibitors. The cellular activity of a lead inhibitor was demonstrated by the inhibition of both PARP and PARG activity in squamous cell carcinoma cells, although preferential inhibition of PARG relative to PARP was observed. The ability of inhibitors to modulate PAR metabolism via simultaneous effects on PARPs and PARG may represent a new approach for therapeutic development.
Figures
Figure 1
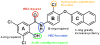
Key pharmacophoric elements related to PARG inhibition
Figure 2
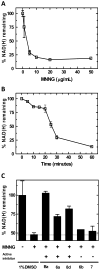
Depletion of NAD(H) in SCC-25 cells following genotoxic stress: (A) Treatment with varying concentrations of MNNG for 30 minutes; (B) A time course of NAD(H) levels following treatment with MNNG (5μg/mL); (C) Effects on NAD(H) depletion in cells treated with MNNG (5μg/mL) for 30 minutes, pretreated with 1mM of benzamide (Bz), 6a, 8d, 6b, or 7.
Figure 3
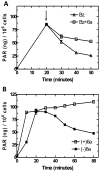
PAR formation in SCC-25 cells following genotoxic stress: (A) Time course of PAR formation after treatment of MNNG (5μg/mL). Cells were treated with 1mM benzamide with compound 6a (□) and without compound 6a (△) at 20 minutes as indicated by the arrow; (B) Time course of PAR formation following treatment of MNNG (5μg/mL), pretreated with 1mM compound 6a (□) and without pretreatment (■).
Scheme 1
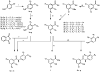
Synthesis of A-ring, B-ring, and C-ring substituted analogues.a _a_Reagents and Conditions: (i) HO-R, Cs2CO3, 90°C, 24 hrs; (ii) SnCl2, EtOH, 70°C, 3 hrs; (iii) CH3I, acetone, K2CO3, reflux, 3 hrs; (iv) SOCl2, 100°C, 30 min; (v) CH2Cl2, RT; (vi) NH4OH, CH2Cl2, RT; (vii) H2N-R, ether, RT.
Scheme 2
Synthesis of linker modified analogues.a _a_Reagents and Conditions: (viii) 3,5-dichloro-salicylaldehyde, EtOH; (ix) NaCNBH3, EtOH; (x) CH3I, acetone, K2CO3, reflux 3 hrs; (xi) 5a, CH2Cl2, RT.
Similar articles
- A high-throughput screening-compatible homogeneous time-resolved fluorescence assay measuring the glycohydrolase activity of human poly(ADP-ribose) glycohydrolase.
Stowell AI, James DI, Waddell ID, Bennett N, Truman C, Hardern IM, Ogilvie DJ. Stowell AI, et al. Anal Biochem. 2016 Jun 15;503:58-64. doi: 10.1016/j.ab.2016.03.016. Epub 2016 Mar 29. Anal Biochem. 2016. PMID: 27036617 - Selective small molecule inhibition of poly(ADP-ribose) glycohydrolase (PARG).
Finch KE, Knezevic CE, Nottbohm AC, Partlow KC, Hergenrother PJ. Finch KE, et al. ACS Chem Biol. 2012 Mar 16;7(3):563-70. doi: 10.1021/cb200506t. Epub 2012 Jan 26. ACS Chem Biol. 2012. PMID: 22220926 Free PMC article. - New Insights into the Roles of NAD+-Poly(ADP-ribose) Metabolism and Poly(ADP-ribose) Glycohydrolase.
Tanuma S, Sato A, Oyama T, Yoshimori A, Abe H, Uchiumi F. Tanuma S, et al. Curr Protein Pept Sci. 2016;17(7):668-682. doi: 10.2174/1389203717666160419150014. Curr Protein Pept Sci. 2016. PMID: 27817743 Review. - Poly(ADP-Ribose) Glycohydrolase (PARG) vs. Poly(ADP-Ribose) Polymerase (PARP) - Function in Genome Maintenance and Relevance of Inhibitors for Anti-cancer Therapy.
Harrision D, Gravells P, Thompson R, Bryant HE. Harrision D, et al. Front Mol Biosci. 2020 Aug 28;7:191. doi: 10.3389/fmolb.2020.00191. eCollection 2020. Front Mol Biosci. 2020. PMID: 33005627 Free PMC article. Review.
Cited by
- Targeting dePARylation for cancer therapy.
Kassab MA, Yu LL, Yu X. Kassab MA, et al. Cell Biosci. 2020 Jan 29;10:7. doi: 10.1186/s13578-020-0375-y. eCollection 2020. Cell Biosci. 2020. PMID: 32010441 Free PMC article. Review. - The role of dePARylation in DNA damage repair and cancer suppression.
Kassab MA, Yu X. Kassab MA, et al. DNA Repair (Amst). 2019 Apr;76:20-29. doi: 10.1016/j.dnarep.2019.02.002. Epub 2019 Feb 8. DNA Repair (Amst). 2019. PMID: 30807923 Free PMC article. Review. - Readers of poly(ADP-ribose): designed to be fit for purpose.
Teloni F, Altmeyer M. Teloni F, et al. Nucleic Acids Res. 2016 Feb 18;44(3):993-1006. doi: 10.1093/nar/gkv1383. Epub 2015 Dec 15. Nucleic Acids Res. 2016. PMID: 26673700 Free PMC article. Review. - Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
Schuller M, Riedel K, Gibbs-Seymour I, Uth K, Sieg C, Gehring AP, Ahel I, Bracher F, Kessler BM, Elkins JM, Knapp S. Schuller M, et al. ACS Chem Biol. 2017 Nov 17;12(11):2866-2874. doi: 10.1021/acschembio.7b00445. Epub 2017 Oct 19. ACS Chem Biol. 2017. PMID: 28991428 Free PMC article. - Antibacterial activity of salicylanilide 4-(trifluoromethyl)-benzoates.
Krátký M, Vinšová J, Novotná E, Mandíková J, Trejtnar F, Stolaková J. Krátký M, et al. Molecules. 2013 Mar 25;18(4):3674-88. doi: 10.3390/molecules18043674. Molecules. 2013. PMID: 23529028 Free PMC article.
References
- Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282(5740):740–741. - PubMed
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35(4):208–219. - PubMed
- Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp Cell Res. 2001;268(1):7–13. - PubMed
- Braun SA, Panzeter PL, Collinge MA, Althaus FR. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur J Biochem. 1994;220(2):369–375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R44 CA090085/CA/NCI NIH HHS/United States
- R01 CA043894/CA/NCI NIH HHS/United States
- R01 CA043894-24/CA/NCI NIH HHS/United States
- CA43894/CA/NCI NIH HHS/United States
- P01 CA027502/CA/NCI NIH HHS/United States
- CA090085/CA/NCI NIH HHS/United States
- CA27502/CA/NCI NIH HHS/United States
- R43 CA090085/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous