ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis - PubMed (original) (raw)
ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis
Jiro Kato et al. Cancer Res. 2011.
Abstract
Protein ADP-ribosylation is a reversible posttranslational modification of uncertain significance in cancer. In this study, we evaluated the consequences for cancer susceptibility in the mouse of a genetic deletion of the enzyme responsible for removing mono-ADP-ribose moieties from arginines in cellular proteins. Specifically, we analyzed cancer susceptibility in animals lacking the ADP-ribosylarginine hydrolase (ARH1) that cleaves the ADP ribose-protein bond. ARH1(-/-) cells or ARH1(-/-) cells overexpressing an inactive mutant ARH1 protein (ARH1(-/-)+dm) had higher proliferation rates than either wild-type ARH1(+/+) cells or ARH1(-/-) cells engineered to express the wild-type ARH1 enzyme. More significantly, ARH1(-/-) and ARH1(+/-) mice spontaneously developed lymphomas, adenocarcinomas, and metastases more frequently than wild-type ARH1(+/+) mice. In ARH1(+/-) mice, we documented in all arising tumors mutation of the remaining wild-type allele (or loss of heterozygosity), illustrating the strict correlation that existed between tumor formation and absence of ARH1 gene function. Our findings show that proper control of protein ADP-ribosylation levels affected by ARH1 is essential for cancer suppression.
Conflict of interest statement
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figures
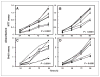
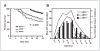
Figure 1
Effect of ARH1 genotype in proliferation of MEFs in vitro. A, ARH1+/+ (■, □), ARH1+/− (▲, △), and _ARH1_−/− (●, ○, ◆) cells (5 × 103) were seeded in 96-well plates and MTT assays were done after growth for the indicated time. B, _ARH1_−/− mock (◇, ◆), _APH1_−/− +dm (▲, △), and _ARH1_−/− +wt (■, □) cells were treated and assayed as in (A). C, ARH1+/+ (■, □), ARH1+/− (▲, △), or _ARH1_−/− (●, ○, ◆) cells were treated as in (A) before BrdU cell proliferation assays. D, _ARH1_−/− mock (◇, ◆), _APH1_−/− +dm (▲, △), or _ARH1_−/− +wt (■, □) cells were subjected to BrdU assays as in (C). Open and closed symbols represent 2 or 3 different cell lines. Data are means ± SEM of values from 8 assays, 3 experiments. Pairwise comparison showed that all genotypes were significantly different from ARH1+/+ or _ARH1_−/− +wt (all at P < 0.0001).
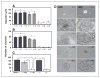
Figure 2
Effect of ARH1 genotype on growth of MEFs in soft agar. A, diameters of colonies _ARH1_−/− (black bars), ARH1+/− (gray bars), and ARH1+/+ (open bars) colonies in soft agar after incubation for 32 days (37°C, with 5% CO2). Data are means ± SEM of values from triplicate assays, 3 experiments of each cell line for colony formation in soft agar. Pairwise comparison showed that _ARH1_−/− and ARH1+/− cells were not different but both were different from ARH1+/+ (*, P < 0.0001). B, numbers of _ARH1_−/− (black bars), _ARH1_+/− (gray bars), and _ARH1_+/+ (open bars) colonies in soft agar after 32 days (37°C, with 5% CO2). Data are means ± SEM of values from triplicate assays, 3 experiments. Pairwise comparison showed that _ARH1_−/− and _ARH1_+/− cells were not different but both were different from _ARH1_+/+ cells (*, _P_ < 0.0001). C, numbers of _ARH1_−/− _mock_ (black bar), _ARH1_−/− +_dm_ (gray bar), _ARH_−/− (dark gray bar), _ARH1_+/+ (open bar), and _ARH1_+/++_wt_ (open bar) colonies (all > 100 μm diameter) in soft agar after 32 days. Data are means ± SEM of values from 3 experiments (triplicate assays) for each cell line. Pairwise comparison showed that _ARH1_−/− mock, _ARH1_−/−, and _ARH1_−/− +dm cells were different from ARH1+/++wt or ARH1+/+ (all at *, P < 0.0001). D, appearance (×400 or ×600) of colonies of indicated genotypes in soft agar after 32 days.
Figure 3
Effect of ARH1 genotype on tumor formation by MEFs in nude mice. A, ARH1+/+ (■, □), ARH1+/− (▲, △), or _ARH1_−/− (●, ○) cells (1 × 106) were injected subcutaneously in nude mice; width plus length of tumor masses was measured 3 times per week thereafter. Data are means ± SEM of values from 5 mice. These experiments were replicated 3 times. Genotypes were significant predictors of maximum volume (P < 0.0001). Pairwise comparisons indicate that _ARH1_−/− and ARH1+/− were different from ARH1+/+ (all at P < 0.0001). B, _ARH1_−/− +wt (■, □), _ARH1_−/− + dm (▲, △), or _ARH1_−/− mock (◆, ◇) cells (1 × 106) were injected subcutaneously in nude mice. Width plus length of tumor masses was measured 3 times per week thereafter. Open and closed symbols represent 2 different cell lines. Data are means ± SEM of values from 5 mice of each group in 3 experiments. Groups of _ARH1_−/− mock and _ARH1_−/− + dm were significantly different from _ARH1_−/− +wt (all at P < 0.0001). C, histology of tumors in nude mice injected with _ARH1_−/−, ARH+/−, _ARH1_−/− mock, and _ARH1_−/− +dm cells [hematoxylin and eosin (H&E) stain].
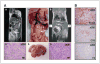
Figure 4
Lymphomas in _ARH1_−/− mice. A (left top), MRI shows tumor mass (lymph node) visible (arrow) in saggital section of 6.5-month _ARH1_−/− mouse. Below is histology (H&E stain, ×200) of lymphoma in GI lymph node. Findings were similar in 2 MRI experiments. Abdominal lymphoma at necropsy in _ARH1_−/− mouse (center top), with metastatic lymphoma excised from mouse liver below. At top right, metastatic lymphoma was visible in liver (arrows) on MRI (frontal section) of 6.5-month _ARH1_−/− mouse. Histology (H&E stain, ×400) of tumor originated in the GI tract of metastatic lymphoma in mouse liver (shown above right). Images in (A-a) were from a different _ARH1_−/− mouse then images designated (A-b). B, immunoreactivity of lymphoma from _ARH1_−/− mouse (×200). B-cell, B-cell marker (top); CD3, T-cell marker (center); F4, macrophage marker (bottom). Experiments were repeated with lymphomas from 8 _ARH1_−/− mice with duplicate analysis in each mouse.
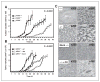
Figure 5
Effect of genotype on survival and incidence of tumors in ARH1-deficient mice. A, effect of genotype on survival of ARH1-deficient mice. Survival data from 91 ARH1+/+ (■), 99 ARH1+/− (◇), and 98 _ARH1_−/− mice (▽) followed 24 months. Every mouse developed some form of tumor and died spontaneously or had to be euthanized due to ill health before 24 months. Pairwise comparisons showed that _ARH1_−/− and ARH1+/− were different from ARH1+/+ (both at *, P < 0.0001). ARH1+/− was not different from _ARH1_−/− (P = 0.093). B, effect of genotype on incidence of tumor in ARH1-deficient mice. Histogram generated after MRI scanning of all ARH1 genotype mice for tumor detection. 490 ARH1+/+ mice, 653 ARH1+/− mice, and 529 _ARH1_−/− mice were subjected to MRI scanning and necropsy at 20 months. ARH1+/+ (open bars), ARH1+/− (gray bars), and _ARH1_−/− (black bars) indicated the number of mice with tumor (left Y axis). ARH1+/+ (□), ARH1+/− (△), and _ARH1_−/− (○) indicated the percentages of mice with tumor (right Y axis). Comparing the cumulative number of tumors by genotype we obtained at P < 0.0001. All pairwise comparisons were significant at P < 0.0001. _ARH1_−/− mice did not differ from ARH1+/− mice in terms of the time to first tumor appearance (P = 0.686).
Figure 6
ARH1 in tumors from ARH1+/− mice. A, immunoblotting with anti-ARH1 antibodies of proteins in lymph node lysates (50 μg) from ARH1_−/−, +/−_, +/+ mouse, and lymphoma from lung (50 μg) of ARH1+/− mouse. Arrows indicate 39-kDa ARH1 protein. Below is immunoreactive α-tubulin on the same blot. Arrows indicate 50-kDa α-tubulin. B, immunoblotting as in A of ARH1 in lysates (80 μg) of lung from ARH1_−/−, +/_3, +/+ mouse, and adenocarcinoma from lung (80 μg) of ARH1+/− mouse. Below is immunoreactive α-tubulin on the same blot.
Similar articles
- Enhanced sensitivity to cholera toxin in female ADP-ribosylarginine hydrolase (ARH1)-deficient mice.
Watanabe K, Kato J, Zhu J, Oda H, Ishiwata-Endo H, Moss J. Watanabe K, et al. PLoS One. 2018 Nov 30;13(11):e0207693. doi: 10.1371/journal.pone.0207693. eCollection 2018. PLoS One. 2018. PMID: 30500844 Free PMC article. - Mutations of the functional ARH1 allele in tumors from ARH1 heterozygous mice and cells affect ARH1 catalytic activity, cell proliferation and tumorigenesis.
Kato J, Vekhter D, Heath J, Zhu J, Barbieri JT, Moss J. Kato J, et al. Oncogenesis. 2015 Jun 1;4(6):e151. doi: 10.1038/oncsis.2015.5. Oncogenesis. 2015. PMID: 26029825 Free PMC article. - Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases.
Mashimo M, Kato J, Moss J. Mashimo M, et al. DNA Repair (Amst). 2014 Nov;23:88-94. doi: 10.1016/j.dnarep.2014.03.005. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746921 Free PMC article. Review. - ARH1 in Health and Disease.
Ishiwata-Endo H, Kato J, Stevens LA, Moss J. Ishiwata-Endo H, et al. Cancers (Basel). 2020 Feb 19;12(2):479. doi: 10.3390/cancers12020479. Cancers (Basel). 2020. PMID: 32092898 Free PMC article. Review. - Understanding the differences of the ligand binding/unbinding pathways between phosphorylated and non-phosphorylated ARH1 using molecular dynamics simulations.
Zhu J, Lv Y, Han X, Xu D, Han W. Zhu J, et al. Sci Rep. 2017 Sep 29;7(1):12439. doi: 10.1038/s41598-017-12031-0. Sci Rep. 2017. PMID: 28963484 Free PMC article.
Cited by
- Uncovering the Invisible: Mono-ADP-ribosylation Moved into the Spotlight.
Hopp AK, Hottiger MO. Hopp AK, et al. Cells. 2021 Mar 19;10(3):680. doi: 10.3390/cells10030680. Cells. 2021. PMID: 33808662 Free PMC article. Review. - Emerging roles of ADP-ribosyl-acceptor hydrolases (ARHs) in tumorigenesis and cell death pathways.
Bu X, Kato J, Moss J. Bu X, et al. Biochem Pharmacol. 2019 Sep;167:44-49. doi: 10.1016/j.bcp.2018.09.028. Epub 2018 Sep 27. Biochem Pharmacol. 2019. PMID: 30267646 Free PMC article. Review. - Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity.
Brady PN, Goel A, Johnson MA. Brady PN, et al. Microbiol Mol Biol Rev. 2018 Dec 19;83(1):e00038-18. doi: 10.1128/MMBR.00038-18. Print 2019 Mar. Microbiol Mol Biol Rev. 2018. PMID: 30567936 Free PMC article. Review. - The NAD metabolome--a key determinant of cancer cell biology.
Chiarugi A, Dölle C, Felici R, Ziegler M. Chiarugi A, et al. Nat Rev Cancer. 2012 Nov;12(11):741-52. doi: 10.1038/nrc3340. Epub 2012 Sep 28. Nat Rev Cancer. 2012. PMID: 23018234 Review. - The cardiac-restricted protein ADP-ribosylhydrolase-like 1 is essential for heart chamber outgrowth and acts on muscle actin filament assembly.
Smith SJ, Towers N, Saldanha JW, Shang CA, Mahmood SR, Taylor WR, Mohun TJ. Smith SJ, et al. Dev Biol. 2016 Aug 15;416(2):373-88. doi: 10.1016/j.ydbio.2016.05.006. Epub 2016 May 20. Dev Biol. 2016. PMID: 27217161 Free PMC article.
References
- Williamson KC, Moss J. Mono-ADP-ribosyltransferases and ADP-ribosylarginine hydrolase: a mono-ADP-ribosylation cycle in animal cells. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, D.C: ASM Press; 1990. pp. 493–510.
- Okazaki IJ, Moss J. Characterization of glycosylphosphatidylinositiol-anchored, secreted, and intracellular vertebrate mono-ADP-ribosyl-transferases. Annu Rev Nutr. 1999;19:485–509. - PubMed
- Moss J, Vaughan M. ADP-ribosylation of guanyl nucleotide-binding regulatory proteins by bacterial toxins. Adv Enzymol Relat Areas Mol Biol. 1988;61:303–79. - PubMed
- Ui M. Pertussis toxin as a valuable probe for G-protein involvement in signal transduction. In: Moss J, Vaughan M, editors. ADP-ribosylating toxins and G proteins: insights into signal transduction. Washington, D.C: ASM Press; 1990. pp. 45–77.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases