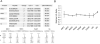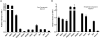Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions - PubMed (original) (raw)
. 2011 Aug 5;10(8):3652-9.
doi: 10.1021/pr2002758. Epub 2011 Jul 8.
Affiliations
- PMID: 21699228
- PMCID: PMC4226403
- DOI: 10.1021/pr2002758
Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions
Chen Ding et al. J Proteome Res. 2011.
Abstract
Cohesin is a protein complex that plays an essential role in pairing replicated sister chromatids during cell division. The vertebrate cohesin complex consists of four core components including structure maintenance of chromosomes proteins SMC1 and SMC3, RAD21, and SA2/SA1. Extensive research suggests that cohesin traps the sister chromatids by a V-shaped SMC1/SMC3 heterodimer bound to the RAD21 protein that closes the ring. Accordingly, the single "ring" model proposes that two sister chromatids are trapped in a single ring that is composed of one molecule each of the 4 subunits. However, evidence also exists for alternative models. The hand-cuff model suggests that each sister chromatid is trapped individually by two rings that are joined through the shared SA1/SA2 subunit. We report here the determination of cohesin subunit stoichiometry of endogenous cohesin complex by quantitative mass spectrometry. Using qConCAT-based isotope labeling, we show that the cohesin core complex contains equimolar of the 4 core components, suggesting that each cohesin ring is closed by one SA1/SA2 molecule. Furthermore, we applied this strategy to quantify post-translational modification-dependent cohesin interactions. We demonstrate that quantitative mass spectrometry is a powerful tool for measuring stoichiometry of endogenous protein core complex.
Figures
Figure 1. Optimal workflow for selection of tryptic peptides for the qConCAT standard
To select best peptides for the qConCAT standard, the immunoprecipitated (IP) cohesin complexes were digested and measured by MS. In addition to the accepted physical and chemical characters, an ideal qConCAT peptide should have (1) high spectral counts (SPCs) in an IP, (2) a linear response curve with the best sensitivity to concentration changes, and (3) show shortest length of time for the complete tryptic digestion in-solution. Two qConCAT peptides are selected for each protein complex subunit. These are fused in tandem to design a qConCAT standard in silico and reverse-translated. The qConCAT gene was chemically synthesized, sub-cloned in an expression vector, and expressed in E.coli cultured in glutamine-free DMEM medium containing heavy lysine and arginine. The immunoprecipitated protein complex and heavy isotope labeled qConCAT are combined in same tube for in-solution digestion. Each pair of analyte peptide and qConCAT peptide is quantified using mass spectrometry.
Figure 2. Optimization and characters of the peptides selected for the cohesin complex qConCAT standard
(A) Labeling efficiency of heavy isotope- qConCAT standard is more than 99%. (B) Validation of stoichiometry measurement of light and heavy labeled qConCAT protein. Equal amount of H and L qConCAT proteins were mixed and the relative ratio of the H/L values were measured by comparing their corresponding peak area of heavy and light isotope labeled qConCAT peptides. All H/L ratios were normalized to the SMC1. The error range represents measurements for the two qConCAT peptides for each of the cohesin proteins.
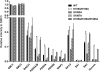
Figure 3. Cohesin complex stoichiometry in SMC1 and PDS5A immunoprecipitations
The cohesin complexes were immunoprecipitated from the MCF7 whole cell extracts and quantified against the qConCATv3 standard. Protein amounts in the two IPs were normalized to their respective antigens. (A) IP of the core cohesin component SMC1 shows potential displacement of RAD21 and SA1/2. (B) IP of the peripheral cohesin component PDS5A, where stoichiometry of the core cohesin complex (SMC1, SMC3, RAD21, and SA1/2) appears equimolar. Furthermore, PDS5A cohesins do not contain PDS5B, and ~20% of cohesin complexes contain SA1, whereas the other ~80% contains SA2.
Figure 4. qConCAT quantification reveals changes in protein associations of the cohesin complexes containing PTM-deficient mutants of SMC3
Flag-SMC3-WT and PTM-deficient SMC3 mutant were immunoprecipitated with anti-Flag M2 Sepharose and quantified against the qConCATstandard. The amount of each complex component was normalized to that of SMC3.
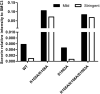
Figure 5. Comparison of relative amount of SORORIN with two washing conditions from wild type or PTM defective SMC3 immunoprecipitations
Flag-SMC3-WT and PTM-deficient SMC3 mutants were immunoprecipitated and washed by two buffers with different detergent contents to dissociate SORORIN from cohesion immunoprecipitates. The relative amount of SORORIN protein level is normalized to that of SMC3.
Similar articles
- Characterization of the interaction between the cohesin subunits Rad21 and SA1/2.
Zhang N, Jiang Y, Mao Q, Demeler B, Tao YJ, Pati D. Zhang N, et al. PLoS One. 2013 Jul 12;8(7):e69458. doi: 10.1371/journal.pone.0069458. Print 2013. PLoS One. 2013. PMID: 23874961 Free PMC article. - Calpain-1 cleaves Rad21 to promote sister chromatid separation.
Panigrahi AK, Zhang N, Mao Q, Pati D. Panigrahi AK, et al. Mol Cell Biol. 2011 Nov;31(21):4335-47. doi: 10.1128/MCB.06075-11. Epub 2011 Aug 29. Mol Cell Biol. 2011. PMID: 21876002 Free PMC article. - A handcuff model for the cohesin complex.
Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D. Zhang N, et al. J Cell Biol. 2008 Dec 15;183(6):1019-31. doi: 10.1083/jcb.200801157. J Cell Biol. 2008. PMID: 19075111 Free PMC article. - The torments of the cohesin ring.
Chavda AP, Ang K, Ivanov D. Chavda AP, et al. Nucleus. 2017 May 4;8(3):261-267. doi: 10.1080/19491034.2017.1295200. Epub 2017 Feb 27. Nucleus. 2017. PMID: 28453390 Free PMC article. Review. - The expanding phenotypes of cohesinopathies: one ring to rule them all!
Piché J, Van Vliet PP, Pucéat M, Andelfinger G. Piché J, et al. Cell Cycle. 2019 Nov;18(21):2828-2848. doi: 10.1080/15384101.2019.1658476. Epub 2019 Sep 13. Cell Cycle. 2019. PMID: 31516082 Free PMC article. Review.
Cited by
- The physical chemistry of interphase loop extrusion.
Tortora MMC, Fudenberg G. Tortora MMC, et al. bioRxiv [Preprint]. 2024 Aug 24:2024.08.23.609419. doi: 10.1101/2024.08.23.609419. bioRxiv. 2024. PMID: 39229088 Free PMC article. Preprint. - Studying macromolecular complex stoichiometries by peptide-based mass spectrometry.
Wohlgemuth I, Lenz C, Urlaub H. Wohlgemuth I, et al. Proteomics. 2015 Mar;15(5-6):862-79. doi: 10.1002/pmic.201400466. Epub 2015 Feb 6. Proteomics. 2015. PMID: 25546807 Free PMC article. Review. - A reference peptide database for proteome quantification based on experimental mass spectrum response curves.
Liu W, Wei L, Sun J, Feng J, Guo G, Liang L, Fu T, Liu M, Li K, Huang Y, Zhu W, Zhen B, Wang Y, Ding C, Qin J. Liu W, et al. Bioinformatics. 2018 Aug 15;34(16):2766-2772. doi: 10.1093/bioinformatics/bty201. Bioinformatics. 2018. PMID: 29617941 Free PMC article. - Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform.
Fernandez-Martinez J, Kim SJ, Shi Y, Upla P, Pellarin R, Gagnon M, Chemmama IE, Wang J, Nudelman I, Zhang W, Williams R, Rice WJ, Stokes DL, Zenklusen D, Chait BT, Sali A, Rout MP. Fernandez-Martinez J, et al. Cell. 2016 Nov 17;167(5):1215-1228.e25. doi: 10.1016/j.cell.2016.10.028. Epub 2016 Nov 10. Cell. 2016. PMID: 27839866 Free PMC article. - Application of targeted mass spectrometry in bottom-up proteomics for systems biology research.
Manes NP, Nita-Lazar A. Manes NP, et al. J Proteomics. 2018 Oct 30;189:75-90. doi: 10.1016/j.jprot.2018.02.008. Epub 2018 Feb 13. J Proteomics. 2018. PMID: 29452276 Free PMC article. Review.
References
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. - PubMed
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat Cell Biol. 2000;2:492–499. - PubMed
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. - PubMed
- Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17:630–636. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM080703/GM/NIGMS NIH HHS/United States
- R01 CA84199/CA/NCI NIH HHS/United States
- R01 GM080703/GM/NIGMS NIH HHS/United States
- R01 CA098500/CA/NCI NIH HHS/United States
- R01 CA98500/CA/NCI NIH HHS/United States
- R01 CA098500-04/CA/NCI NIH HHS/United States
- R01 CA084199/CA/NCI NIH HHS/United States
- R01 CA084199-08/CA/NCI NIH HHS/United States
- R01 GM080703-03/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous