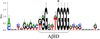The Alzheimer's amyloid β-peptide (Aβ) binds a specific DNA Aβ-interacting domain (AβID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif - PubMed (original) (raw)
The Alzheimer's amyloid β-peptide (Aβ) binds a specific DNA Aβ-interacting domain (AβID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif
Bryan Maloney et al. Gene. 2011.
Abstract
Deposition of extracellular plaques, primarily consisting of amyloid β peptide (Aβ), in the brain is the confirmatory diagnostic of Alzheimer's disease (AD); however, the physiological and pathological role of Aβ is not fully understood. Herein, we demonstrate novel Aβ activity as a putative transcription factor upon AD-associated genes. We used oligomers from 5'-flanking regions of the apolipoprotein E (APOE), Aβ-precursor protein (APP) and β-amyloid site cleaving enzyme-1 (BACE1) genes for electrophoretic mobility shift assay (EMSA) with different fragments of the Aβ peptide. Our results suggest that Aβ bound to an Aβ-interacting domain (AβID) with a consensus of "KGGRKTGGGG". This peptide-DNA interaction was sequence specific, and mutation of the first "G" of the decamer's terminal "GGGG" eliminated peptide-DNA interaction. Furthermore, the cytotoxic Aβ25-35 fragment had greatest DNA affinity. Such specificity of binding suggests that the AβID is worth of further investigation as a site wherein the Aβ peptide may act as a transcription factor.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures
Figure 1. Presence of Aβ–binding motifs (AβID) with 80% homology to p53 HSE sequence on APOE, APP, and BACE1 5’–flanking sequences
The 5’–flanking sequences of the APOE, APP, and BACE1 genes were searched for decamers with at least 80% homology to the “GGATTGGGGT” Aβ–binding HSE site of the TP53 promoter (Ohyagi et al., 2005). Sites marked with “*” were chosen for further study in this report.
A.
A region of the APOE gene from 2kb upstream of the +1 TSS to the end of the first coding exon, which contains the first intron, (Paik et al., 1985; Du et al., 2005) was searched.
B.
The APP 5’–flanking region from 4kb upstream of the +1 TSS to the end of the first coding exon (Lahiri and Robakis, 1991; Hattori et al., 1997) was searched.
C.
The BACE1 5’ flanking region from 3.8kb upstream of the +1 TSS to the “ATG” start codon (Christensen et al., 2004; Sambamurti et al., 2004) was searched.
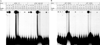
Figure 2. Electrophoretic mobility shift assay (EMSA)/gel shift assay of Aβ1–42 vs. different putative binding decamer–containing oligomers
Figure shows representative EMSA of the
A.
Aβ1–42 and the
B,
Aβ1–40 peptides. The reverse Aβ40–1 and 42–1 peptides were independently incubated with radiolabeled oligomer containing the putative AβID at −3833 (lane 1). Aβ1–42 or 1–40 (lanes 2–16) were incubated with radiolabeled oligomers that contained the putative AβIDs in the APP (lanes 2–8) 5’–flanking region at positions −3833G (lane 2), −3833A with a “G” to “A” substitution at −3829 (lane 3), −3364 (lane 4), −2871 (lane 5) and −1862 (lane 6). In addition, an oligomer corresponding to the FAD mutation site in APP at −1023 (lane 7) and a distinct HSE–binding site within APP (lane 8) were incubated and run; in the APOE (lanes 9–12) 5’–flanking region at positions −899 (lane 9), +171 (lane 10), +284 (lane 11), +660/+665 (lane 12); and in the BACE1 (lanes 13–16) 5’–flanking region at positions −1939 (lane 13), −1766 (lane 14), −119 (lane 15), and +36 (lane 16). The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near the top of the gel, unbound oligomer ran to the bottom of the gel.
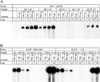
Figure 3. Determination of “binding” vs. “non–binding oligomer pairs to Aβ peptides in EMSA
EMSA films were densitometrically scanned and signals within each film were normalized by subtracting the mean of that signal’s film from the individual signal and dividing by the standard deviation of signals. Normalized signals were then analyzed by Waller–Duncan means separation, and categories indicated with letters on the figure. Samples sharing the same letter were not significantly different at k = 100 (analogous to α = 0.05). The two “background control” oligomer pairs, _APP_HSE and _APP_−1023, indicated by “*”.
Figure 4. Competition EMSA with “positive” oligomer pairs from APOE, APP, BACE1 and TP53 promoters vs. Aβ1–42 peptide
A.
Aβ1–42 peptide was incubated with radiolabeled oligomers that contained the putative AβIDs in the APOE (lanes 1–4) 5’–flanking region at positions +171 (lane 1), −899 (lane 2), +284 (lane 3), +660/+665 (lane 4); in the APP (lanes 5–8) 5’–flanking region at positions −1862 (lane 5), −2871 (lane 6), −3364 (lane 7), and −3833 (lane 8); and in the BACE1 (lanes 9–12) 5’–flanking region at positions −119 (lane 9), −1766 (lane 10), −1939 (lane 11), and +36 (lane 12). In addition, an oligomer containing the Aβ–binding element from p53 was labeled and run (lane 13). The gel was dried and exposed to X–ray film for autoradiography. Specific oligomer pairs that were selected for competition EMSA are indicated with “*”.
B.
Aβ1–42 was incubated with radiolabeled oligomers that had previously shown interaction with Aβ1–42 (lanes 1,3,5,7,9,11,13,15). These reactions were repeated with the addition of 140× molar excess unlabeled oligomer (lanes 2,4,6,8,10,12,14,16). Specificity of Aβ/oligomer interaction is demonstrated by a significant reduction of autoradiograph signal at top of lane. The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near the top of the gel.
Figure 5. Structure of the Aβ–binding domain (AβID) consensus motif
Combined height of stacked letters corresponds to bits of information (Shannon, 1997). An asterisk indicates the position of a “G” that may be critical for Aβ binding activity.
Figure 6. EMSA of different fragments of the Aβ peptide vs. four different DNA probes
Fragments of Aβ peptide corresponding to residues 1–42, 1–40, 1–28, 20–29, 25–35, 29–40, and 31–35 were self–oligomerized. In addition, “reverse” fragments 42–1, 40–1, and 35–25 were prepared.
A.
Fragments were incubated against oligomers the contained the Aβ–binding sequence at APOE +171 (lanes 1–10) or APOE +660 (lanes 11–20). The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near the top of the gel.
B.
Fragments were incubated against oligomers the contained the Aβ–binding sequence at APP −3833G (lanes 1–10) or APP −3833A (with a “G”→“A” substitution) (lanes 11–20). The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near the top of the gel.
Figure 7. Sequences and alignment of different Aβ fragments used in EMSA
All diagrams are aligned and to the same scale.
A.
“Helical–form” secondary structure of Aβ1–42 in aqueous environment (Tomaselli et al., 2006), helices indicated by cylinders.
B.
Secondary structure of Aβ1–42 in apolar solvent (Crescenzi et al., 2002), helices indicated by cylinders.
C.
Forward sequences. Sequences for Aβ 1–42, 1–40, 1–28, 20–29, 25–35, 29–40, and 31–35 are depicted as aligned amino acid residues. Those peptides with no apparent DNA–binding activity are in gray background.
D.
Reverse sequences. Sequences for Aβ42–1, 40–1, and 35–25 are depicted as aligned amino acid residues. Those peptides with no apparent DNA–binding activity are in gray background.
Figure 8. Concentration dependency of Aβ–DNA interaction
A.
The oligomer containing the Aβ–binding consensus at APP −3833G was incubated with fragments of the Aβ peptide, specifically Aβ1–40 (lanes 2–6), 1–42 (lanes 7–10), 42–1 (lanes 11–13), 1–28 (lanes 14–16), and 25–35 (lanes 17–20) at different peptide concentrations as indicated. The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near top of gel.
B.
The oligomers containing the Aβ–binding consensus at APOE +660 (lanes 1–10) and at BACE1 −119 (lanes 11–20) were incubated with fragments of the Aβ peptide, specifically Aβ1–40 (lanes 1–2, 11–12), 1–42 (lanes 3–4, 13–14), 42–1 (lanes 5–6, 15–16), 1–28 (lanes 7–8, 17–18), and 25–35 (lanes 9–10, 19–20) at different peptide concentrations as indicated. The gel was dried and exposed to X–ray film for autoradiography. DNA–peptide interactions appeared as dark signal near top of gel.
Figure 9. Presence of confirmed AβIDs and putative motifs as predicted by dynamic weight scores on selected gene 5’−flanking regions
The 5’–flanking sequences of several genes were searched for decamers, using the weight matrix we generated. Matrix–matching sites are indicated on the sequences. Forward–orientation sites are above the sequence line. Reverse–orientation sites are below the sequence line. The 5’–flanking regions from 4kb upstream to 1 kb downstream of the +1 transcription start site were analyzed for the APOE, APP, BACE1, TP53, ASCL1, BACE2, IDE, MAPT, OLIG2, and SLC38A1 gene sequences. Positions that corresponded to oligomers for our EMSA herein, all positive on EMSA, are boldface. A site on the APP sequence marked with “§” crosses a previously–characterized APP polymorphism associated with late–onset familial AD.
Similar articles
- Alternative Selection of β-Site APP-Cleaving Enzyme 1 (BACE1) Cleavage Sites in Amyloid β-Protein Precursor (APP) Harboring Protective and Pathogenic Mutations within the Aβ Sequence.
Kimura A, Hata S, Suzuki T. Kimura A, et al. J Biol Chem. 2016 Nov 11;291(46):24041-24053. doi: 10.1074/jbc.M116.744722. Epub 2016 Sep 29. J Biol Chem. 2016. PMID: 27687728 Free PMC article. - Amyloid-β protein (Aβ) Glu11 is the major β-secretase site of β-site amyloid-β precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Aβ Asp1 contributes to Alzheimer pathogenesis.
Deng Y, Wang Z, Wang R, Zhang X, Zhang S, Wu Y, Staufenbiel M, Cai F, Song W. Deng Y, et al. Eur J Neurosci. 2013 Jun;37(12):1962-9. doi: 10.1111/ejn.12235. Eur J Neurosci. 2013. PMID: 23773065 - Alzheimer's disease.
De-Paula VJ, Radanovic M, Diniz BS, Forlenza OV. De-Paula VJ, et al. Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review. - The beta-secretase, BACE: a prime drug target for Alzheimer's disease.
Vassar R. Vassar R. J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
Cited by
- Nuclear translocation uncovers the amyloid peptide Aβ42 as a regulator of gene transcription.
Barucker C, Harmeier A, Weiske J, Fauler B, Albring KF, Prokop S, Hildebrand P, Lurz R, Heppner FL, Huber O, Multhaup G. Barucker C, et al. J Biol Chem. 2014 Jul 18;289(29):20182-91. doi: 10.1074/jbc.M114.564690. Epub 2014 May 30. J Biol Chem. 2014. PMID: 24878959 Free PMC article. - Identification and characterization of the Hfq bacterial amyloid region DNA interactions.
Turbant F, Hamoui OE, Partouche D, Sandt C, Busi F, Wien F, Arluison V. Turbant F, et al. BBA Adv. 2021 Oct 30;1:100029. doi: 10.1016/j.bbadva.2021.100029. eCollection 2021. BBA Adv. 2021. PMID: 37082015 Free PMC article. - Chronic Cerebral Hypoperfusion Promotes Amyloid-Beta Pathogenesis via Activating β/γ-Secretases.
Cai Z, Liu Z, Xiao M, Wang C, Tian F. Cai Z, et al. Neurochem Res. 2017 Dec;42(12):3446-3455. doi: 10.1007/s11064-017-2391-9. Epub 2017 Aug 24. Neurochem Res. 2017. PMID: 28836062 - Calmodulin and Amyloid Beta as Coregulators of Critical Events during the Onset and Progression of Alzheimer's Disease.
O'Day DH. O'Day DH. Int J Mol Sci. 2023 Jan 11;24(2):1393. doi: 10.3390/ijms24021393. Int J Mol Sci. 2023. PMID: 36674908 Free PMC article. - Icariin Ameliorates Amyloid Pathologies by Maintaining Homeostasis of Autophagic Systems in Aβ1-42-Injected Rats.
Jiang X, Chen LL, Lan Z, Xiong F, Xu X, Yin YY, Li P, Wang P. Jiang X, et al. Neurochem Res. 2019 Dec;44(12):2708-2722. doi: 10.1007/s11064-019-02889-z. Epub 2019 Oct 15. Neurochem Res. 2019. PMID: 31612304
References
- Barrantes A, Rejas MT, Benitez MJ, Jimenez JS. Interaction between Alzheimer's Abeta1-42 peptide and DNA detected by surface plasmon resonance. J. Alzheimers. Dis. 2007;12:345–355. - PubMed
- Baruch-Suchodolsky R, Fischer B. Abeta40, either soluble or aggregated, is a remarkably potent antioxidant in cell-free oxidative systems. Biochemistry. 2009;48:4354–4370. - PubMed
- Bogoyevitch MA, Boehm I, Oakley A, Ketterman AJ, Barr RK. Targeting the JNK MAPK cascade for inhibition: basic science and therapeutic potential. Biochim. Biophys Acta. 2004;1697:89–101. - PubMed
- Checler F, Dunys J, Pardossi-Piquard R, Alves da Costa C. p53 is regulated by and regulates members of the gamma-secretase complex. Neurodegener. Dis. 2010;7:50–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AG18379/AG/NIA NIH HHS/United States
- R01 AG018884/AG/NIA NIH HHS/United States
- AG18884/AG/NIA NIH HHS/United States
- R03 AG014882/AG/NIA NIH HHS/United States
- R01 AG018379/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous