Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest - PubMed (original) (raw)
Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest
Ian G Ganley et al. Mol Cell. 2011.
Abstract
Autophagy, a catabolic pathway that delivers cellular components to lysosomes for degradation, can be activated by stressful conditions such as nutrient starvation and endoplasmic reticulum (ER) stress. We report that thapsigargin, an ER stressor widely used to induce autophagy, in fact blocks autophagy. Thapsigargin does not affect autophagosome formation but leads to accumulation of mature autophagosomes by blocking autophagosome fusion with the endocytic system. Strikingly, thapsigargin has no effect on endocytosis-mediated degradation of epidermal growth factor receptor. Molecularly, while both Rab7 and Vps16 are essential regulatory components for endocytic fusion with lysosomes, we found that Rab7 but not Vps16 is required for complete autophagy flux, and that thapsigargin blocks recruitment of Rab7 to autophagosomes. Therefore, autophagosomal-lysosomal fusion must be governed by a distinct molecular mechanism compared to general endocytic fusion.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
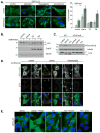
Figure 1. Thapsigargin induces autophagosome accumulation in an ATG5 and phosphatidylinositol 3-kinase activity dependent manner
(A) MEF cells stably expressing GFP-LC3 (green) were incubated in complete media (control), amino acid-free media for 2 h (starvation), complete media with 3 μM thapsigargin for 5 h, or complete media with 20 μg/ml tunicamycin for 5 h; each in the presence or absence of 10 nM bafilomycin A1 for the final two hours of treatment. Cells were stained with DAPI (blue). Bar, 10 μm. Quantitation shown on the right represents mean GFP puncta per cell (n=20) from two independent experiments ± SD. (B) Immunoblots of MEF lysates treated as in (A). (C) Immunoblots of wild type (WT) and ATG5 null MEF lysates, treated as indicated. (D) ATG5 null MEFs, transduced with GFP-ATG5 retrovirus, were treated as indicated, each in the presence or absence of 50 nM wortmannin. Cells were stained with DAPI (blue), anti-GFP (green) and endogenous LC3 (red). Asterisks represent cells that do not express GFP-ATG5, note the lack of LC3 puncta in these cells. Bar, 10 μm. (E) GFP-LC3 expressing MEFs were incubated in amino acid-free media for 1 h (starve), amino acid-free media for 1 h followed by addition of 50 nM wortmannin for 1 h (starve → wort), complete media with 3 μM thapsigargin for 4 h (TG), complete media with 3 μM thapsigargin for 4 h followed by addition of 50 nM wortmannin for 1 h (TG → wort), or complete media with 3 μM thapsigargin and 50 nM wortmannin together for 4 h (TG + wort). Bar, 10 μm. See also Figure S1.
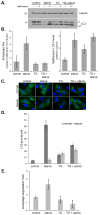
Figure 2. Thapsigargin blocks both basal and starvation-induced autophagy
(A) MEF cells were incubated in complete media (control), amino acid-free media for 1 h (starve), complete media with 3 μM thapsigargin for 5 h (TG), or complete media with 3 μM thapsigargin for 4 h followed by washing and incubation in amino acid-free media with 3 μM thapsigargin for 1 h (TG + starve); each in the presence or absence of 10 nM bafilomycin A1 for the final hour. (B) Quantitation of immunoblot data in (A) showing autophagic flux (change in LC3-II levels upon bafilomycin treatment) and relative bafilomycin-induced LC3-II levels. Values represent means normalized to tubulin from 5 independent experiments ± SEM. (C) GFP-LC3 expressing MEFs were treated as in (A). For rescue samples, cells were cultured in fresh complete media for an additional hour before fixation. Bar, 10 μm. (D) Quantitation of data presented in (C) shows mean number of GFP-LC3 puncta per cell ± SEM (n=50 cells per condition). (E) MEF cells were treated with 3 μM thapsigargin (TG) and/or amino acid-free media (starve) and the increase in degradation of long-lived proteins was measured. Values represent the mean percentage of protein loss per hour ± SD as compared to bafilomycin treated samples from 3 independent experiments.
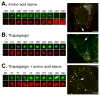
Figure 3. Thapsigargin blocks fusion of autophagosomes with lysosomes
MEF cells stably expressing GFP-mCherry-LC3 were subject to live cell microscopy. Small panels on the left show the life span of an individual LC3 structure, as manifested by its GFP (green, top) and mCherry (red, bottom) fluorescence. The same structure is indicated by an arrow in the whole cell pictured on the right. In (A) times represent minutes post amino acid starvation (see also Movie S1); in (B) times represent minutes post addition of thapsigargin to 3 μM (see also Movie S2); and in (C) times show minutes post amino acid starvation in combination with thapsigargin treatment (see also Movie S3). Bar, 10 μm. See also Figure S2.
Figure 4. Thapsigargin does not block initial stages of autophagosome formation
(A) ATG5 null MEFs stably expressing GFP-ATG5K130R were incubated in complete media (control), amino acid-free media for 1 h (starve), or 3 μM thapsigargin for 4 h in the presence of DMSO or 50 nM wortmannin. Samples were stained with anti-GFP (green) and DAPI (blue). Bar, 10 μm. (B) U2OS cells expressing GFP-LC3 were incubated in complete media (control), amino acid-free media for 2 h (starve), or 3 μM thapsigargin for 5 h, followed by staining with DAPI (blue), anti-p62 (red) and GFP (green). Bar, 10 μm. Panels on the right are high magnifications of the boxed regions; arrows indicate co-localization between LC3 and p62, arrowheads indicate p62-only puncta and double arrowhead indicates LC3-only puncta. (C) Immunoblots of U2OS cell lysates, treated as above. (D) Quantitation of p62 levels shown in (C). Values represent relative levels ± SD, normalized to actin from three independent experiments. See also Figure S3.
Figure 5. Thapsigargin arrests autophagy after autophagosome formation
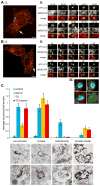
(A) MEFs expressing GFP-LC3 and dsRed-ER were incubated in amino acid-free media and subjected to time-lapse confocal microscopy. (i) An example cell with the arrow marking a single GFP-LC3 puncta whose lifetime is represented in the panels of (ii). Time represents minutes and seconds of amino acid starvation. Bar, 10 μm. See also Movie S4. (B) MEFs expressing GFP-LC3 and dsRed-ER were treated with 3 μM thapsigargin for 4 h followed by amino acid-free media, also containing 3 μM thapsigargin. (i) An example cell with the arrow marking a single GFP-LC3 puncta whose lifetime is represented in the panels of (ii). Time shows minutes of amino acid starvation. (iii) Magnification of the GFP-LC3 puncta from the last panel of (B. ii.) in x, y and z dimensions, plus a 3D reconstruction in the bottom right panel. See also Movie S5. (C) Immuno-EM analysis of GFP-LC3 labeled structures, which were classified into four different groups: non-circular, circular, internal label only, or circular clusters. Bars represent mean number of structures per cell section ± SEM for control treated cells (n = 73 cell sections), 1 h 30 min amino acid-free media treated cells (starve; n = 84 cell sections), 4 h 30 min thapsigargin (3 μM) treated cells (TG; n = 103 cell sections), and 4 h 30 min thapsigargin (3 μM) plus amino acid starvation for the final 1 h 30 min (TG + starve; n = 89 cell sections). Panels below the chart represent examples of the indicated structures. Bar, 500 nm. See also Movie S6.
Figure 6. Thapsigargin does not block endocytosis-mediated EGFR degradation
(A) MEFs were treated as described in the methods section for the EGF receptor degradation assay. Post EGF addition, cells were chased in cycloheximide, with/without 3 μM thapsigargin, or 10 nM bafilomycin A1. (B) Quantitation of EGFR degradation. Values represent mean percentage decrease in EGF receptor levels ± SD from three independent experiments. (C) MEFs were incubated in complete media (control) or complete media with 3 μM thapsigargin for 5 h (thapsigargin). Following 2 h of thapsigargin treatment, 5 mg/ml FITC-dextran (green) was added for a further 2.5 h and then cells were washed and incubated in complete media containing 100 nM Lysotracker (red) with/without thapsigargin for the final 30 min. Cells were stained with DAPI (blue). Bar, 10 μm. (D) Quantitation of data represented in (C). Values represent mean number of dextran structures per cell that were positive for lysotracker stain ± SD, from two independent experiments and a total of 20 cells. See also Figure S4.
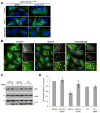
Figure 7. Rab7, but not Vps16, is required for autophagosome fusion with lysosomes
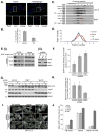
(A) MEF cells were starved for 1 h in amino acid-free media or treated with 3 μM thapsigargin as indicated. Cells stained with DAPI (blue), antibodies against Rab7 (green) and LC3 (red). Shown is a combined confocal image stack. The boxed region is reproduced below the main image displaying a single section highlighting the localization of Rab7 and LC3. Bar, 10 μm. (B) Quantitation of data represented in (A) showing the mean percentage ± SD, from 20 cells in two independent experiments, of LC3 structures co-localizing with Rab7. Rab7 positive structures per cell remained relatively constant with mean values of 38 ± 4 under starvation conditions, compared to 45 ± 9 under thapsigargin treatment. (C) MEF cells were incubated in complete media (control), amino acid-free media for 1 h (starve), complete media with 3 μM thapsigargin for 5 h (TG), or complete media with 3 μM thapsigargin for 5 h amino with acid starvation for the final hour (TG + starve). Cell homogenates were subject to Iodixonal density gradient centrifugation (10%-50% gradient) and fractions immunoblotted. The boxed regions highlight the thapsigargin block of Rab7 increase in fraction 6. (D) Quantitation of the data presented in (C). Values represent mean Rab7 levels in the indicated fractions ± SEM from three independent experiments. (E) EGFR degradation in MEF cells stably expressing control shRNA, Rab 7 shRNA or Vps16 shRNA (the left panel (ii) shows the RNAi efficiency). (F) Quantitation of EGFR degradation. Values represent EGFR levels relative to the start of chase from 4 independent experiments ± SEM. (G) MEF cells stably expressing GFP-LC3 and control shRNA, Rab 7 shRNA or Vps16 shRNA were incubated in complete media or amino acid-free media for 1 h in the presence or absence of 10 nM Bafilomycin A1. (H) Quantitation of the GFP-LC3 immunoblot in (G) showing autophagic flux upon starvation (change in LC3-II levels upon bafilomycin treatment). Values represent means normalized to tubulin from 3 independent experiments ±SEM. (I) MEF cells stably expressing GFP-LC3 and control shRNA, Rab 7 shRNA or Vps16 shRNA were incubated in complete media, amino acid-free media for 1 h (starve) or amino acid-free media for 1 h followed by addition of 50 nM wortmannin for 1h (starve → wort). Bar, 10-μm. (J) Quantitation of data in (I) showing mean number of GFP-LC3 puncta per cell ± SEM (n=20 cells per condition). See also Figure S5.
Similar articles
- Thapsigargin distinguishes membrane fusion in the late stages of endocytosis and autophagy.
Ganley IG, Wong PM, Jiang X. Ganley IG, et al. Autophagy. 2011 Nov;7(11):1397-9. doi: 10.4161/auto.7.11.17651. Epub 2011 Nov 1. Autophagy. 2011. PMID: 21921693 - Cadmium-induced cytotoxicity in mouse liver cells is associated with the disruption of autophagic flux via inhibiting the fusion of autophagosomes and lysosomes.
Zou H, Wang T, Yuan J, Sun J, Yuan Y, Gu J, Liu X, Bian J, Liu Z. Zou H, et al. Toxicol Lett. 2020 Mar 15;321:32-43. doi: 10.1016/j.toxlet.2019.12.019. Epub 2019 Dec 17. Toxicol Lett. 2020. PMID: 31862506 - Cadmium disrupts autophagic flux by inhibiting cytosolic Ca2+-dependent autophagosome-lysosome fusion in primary rat proximal tubular cells.
Liu F, Wang XY, Zhou XP, Liu ZP, Song XB, Wang ZY, Wang L. Liu F, et al. Toxicology. 2017 May 15;383:13-23. doi: 10.1016/j.tox.2017.03.016. Epub 2017 Mar 25. Toxicology. 2017. PMID: 28347754 - Rab7 may be a novel therapeutic target for neurologic diseases as a key regulator in autophagy.
Wen H, Zhan L, Chen S, Long L, Xu E. Wen H, et al. J Neurosci Res. 2017 Oct;95(10):1993-2004. doi: 10.1002/jnr.24034. Epub 2017 Feb 10. J Neurosci Res. 2017. PMID: 28186670 Review. - Hijacking intracellular membranes to feed autophagosomal growth.
Staiano L, Zappa F. Staiano L, et al. FEBS Lett. 2019 Nov;593(22):3120-3134. doi: 10.1002/1873-3468.13637. Epub 2019 Oct 21. FEBS Lett. 2019. PMID: 31603532 Review.
Cited by
- α-Mangostin Reduced ER Stress-mediated Tumor Growth through Autophagy Activation.
Kim SJ, Hong EH, Lee BR, Park MH, Kim JW, Pyun AR, Kim YJ, Chang SY, Chin YW, Ko HJ. Kim SJ, et al. Immune Netw. 2012 Dec;12(6):253-60. doi: 10.4110/in.2012.12.6.253. Epub 2012 Dec 31. Immune Netw. 2012. PMID: 23396851 Free PMC article. - Intracellular cholesterol transport inhibition Impairs autophagy flux by decreasing autophagosome-lysosome fusion.
Maharjan Y, Dutta RK, Son J, Wei X, Park C, Kwon HM, Park R. Maharjan Y, et al. Cell Commun Signal. 2022 Nov 25;20(1):189. doi: 10.1186/s12964-022-00942-z. Cell Commun Signal. 2022. PMID: 36434621 Free PMC article. - Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease.
Lim JA, Li L, Kakhlon O, Myerowitz R, Raben N. Lim JA, et al. Autophagy. 2015;11(2):385-402. doi: 10.1080/15548627.2015.1009779. Autophagy. 2015. PMID: 25758767 Free PMC article. - The neuroprotective effects of glucagon-like peptide 1 in Alzheimer's and Parkinson's disease: An in-depth review.
Reich N, Hölscher C. Reich N, et al. Front Neurosci. 2022 Sep 1;16:970925. doi: 10.3389/fnins.2022.970925. eCollection 2022. Front Neurosci. 2022. PMID: 36117625 Free PMC article. Review. - Sterile activation of invariant natural killer T cells by ER-stressed antigen-presenting cells.
Bedard M, Shrestha D, Priestman DA, Wang Y, Schneider F, Matute JD, Iyer SS, Gileadi U, Prota G, Kandasamy M, Veerapen N, Besra G, Fritzsche M, Zeissig S, Shevchenko A, Christianson JC, Platt FM, Eggeling C, Blumberg RS, Salio M, Cerundolo V. Bedard M, et al. Proc Natl Acad Sci U S A. 2019 Nov 19;116(47):23671-23681. doi: 10.1073/pnas.1910097116. Epub 2019 Nov 5. Proc Natl Acad Sci U S A. 2019. PMID: 31690657 Free PMC article.
References
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. - PubMed
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. - PubMed
- Chan EY. mTORC1 phosphorylates the ULK1-mAtg13-FIP200 autophagy regulatory complex. Sci Signal. 2009;2:pe51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_UP_A500_1019/MRC_/Medical Research Council/United Kingdom
- R01 CA113890/CA/NCI NIH HHS/United States
- R01 CA113890-01A1/CA/NCI NIH HHS/United States
- R01CA113890/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials